Taxa Name
| Taxa
Level | Barcode
Data? | Number of
Barcodes
from
any ocean | Number of
Barcodes
from this region | Barcode Locations and
Species Occurence Map | MZGdb Atlas
v2026-m01-01
2026-Jan-26 |
| Agonus cataphractus |
Species
(1) |
COI
12S
16S
18S
28S
|
COI = 24
12S = 0
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003046
R:1:0:0:0 |
| Ammodytes marinus |
Species
(2) |
COI
12S
16S
18S
28S
|
COI = 50
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 12
12S = 0
16S = 0
18S = 0
28S = 0
|
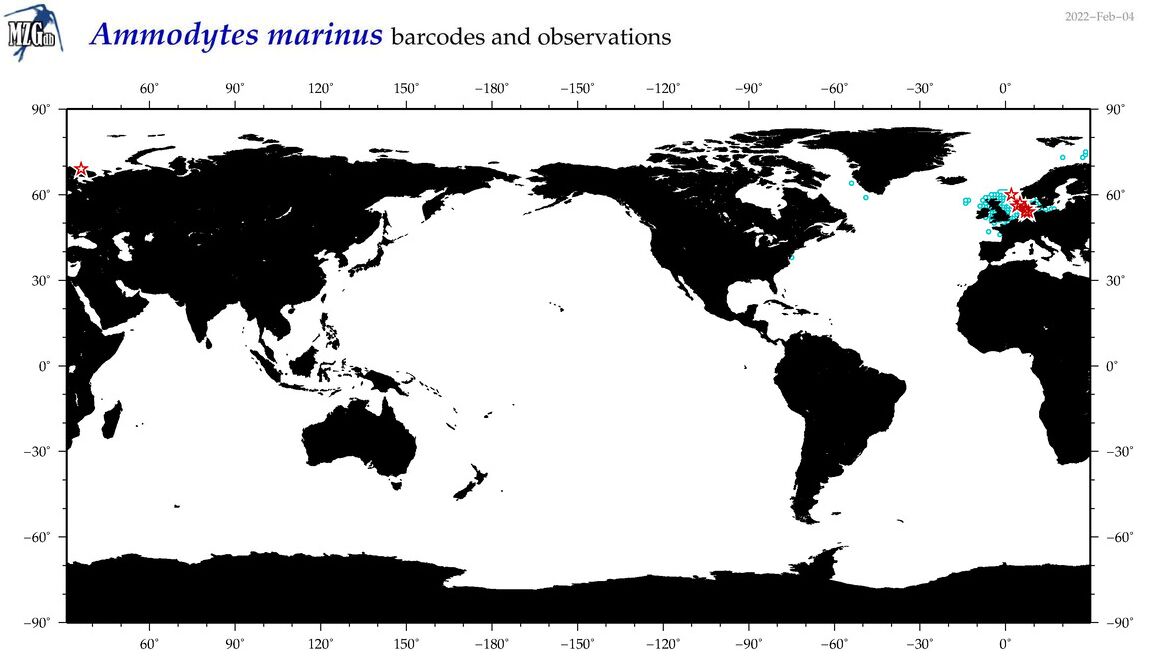 |
accepted
T5016163
R:1:1:0:0 |
| Ammodytes tobianus |
Species
(3) |
COI
12S
16S
18S
28S
|
COI = 8
12S = 2
16S = 2
18S = 2
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
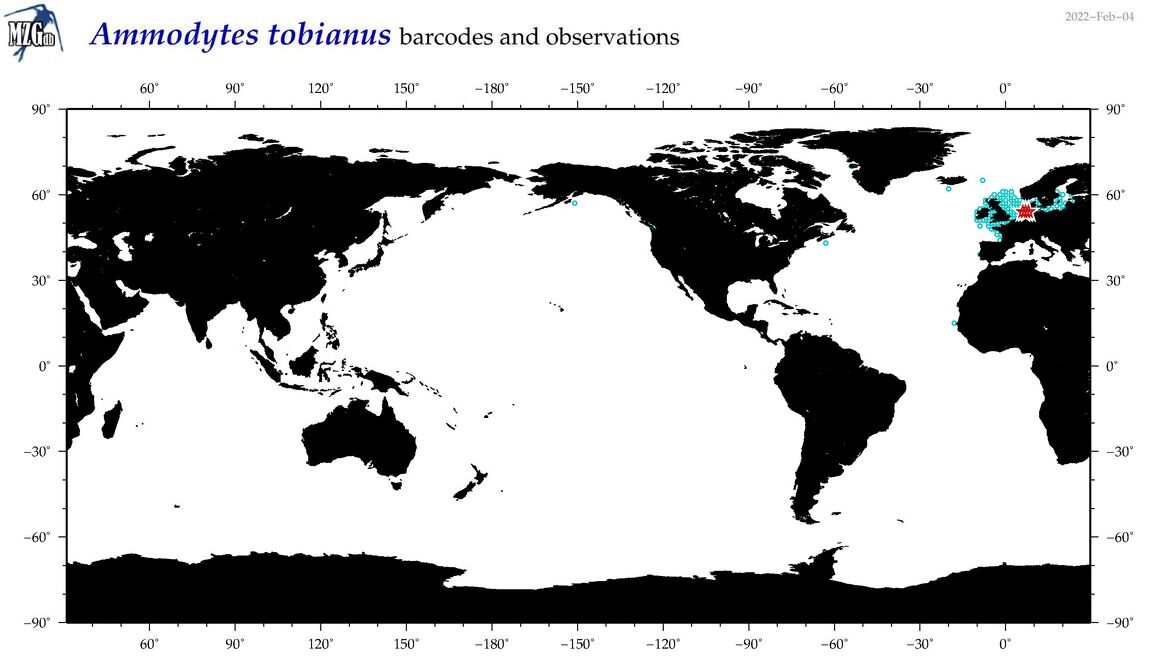 |
accepted
T5016165
R:1:1:0:0 |
| Anarhichas denticulatus |
Species
(4) |
COI
12S
16S
18S
28S
|
COI = 25
12S = 24
16S = 22
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
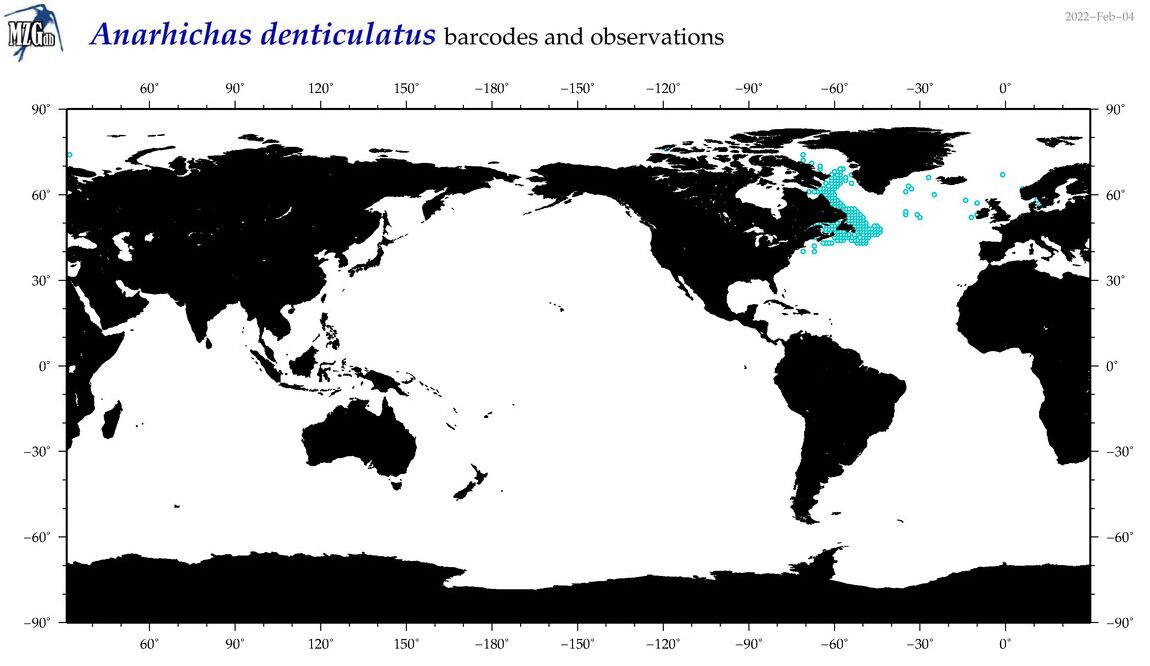 |
accepted
T5003089
R:1:0:0:0 |
| Anarhichas lupus |
Species
(5) |
COI
12S
16S
18S
28S
|
COI = 187
12S = 90
16S = 91
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
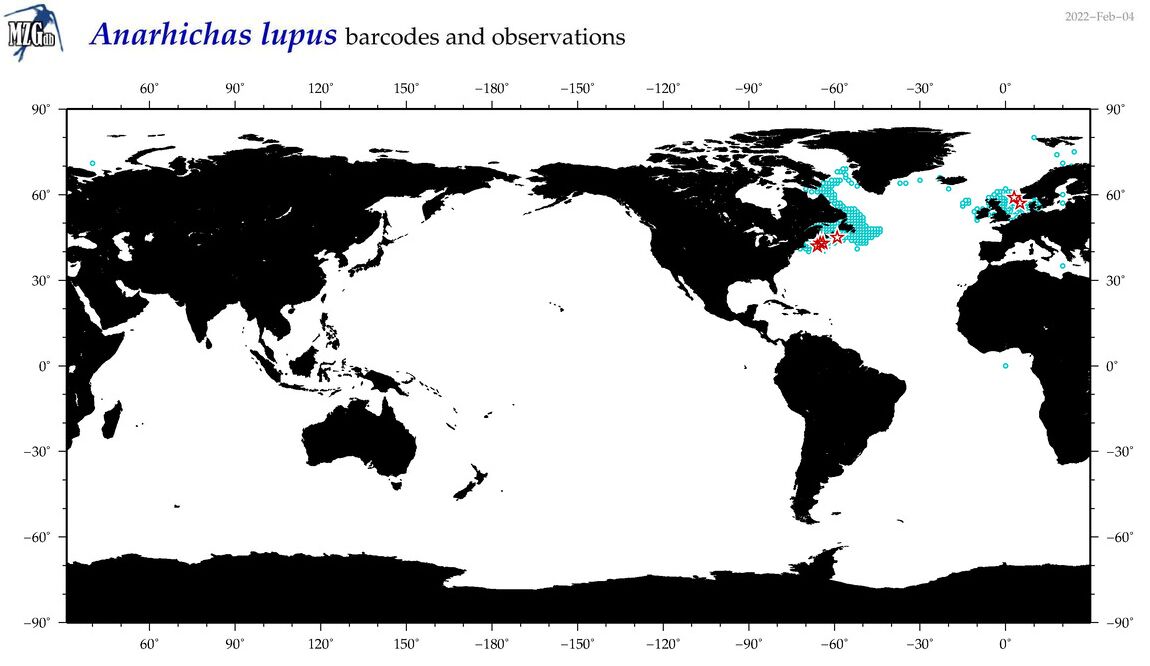 |
accepted
T5003090
R:1:0:0:0 |
| Anarhichas minor |
Species
(6) |
COI
12S
16S
18S
28S
|
COI = 21
12S = 20
16S = 19
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
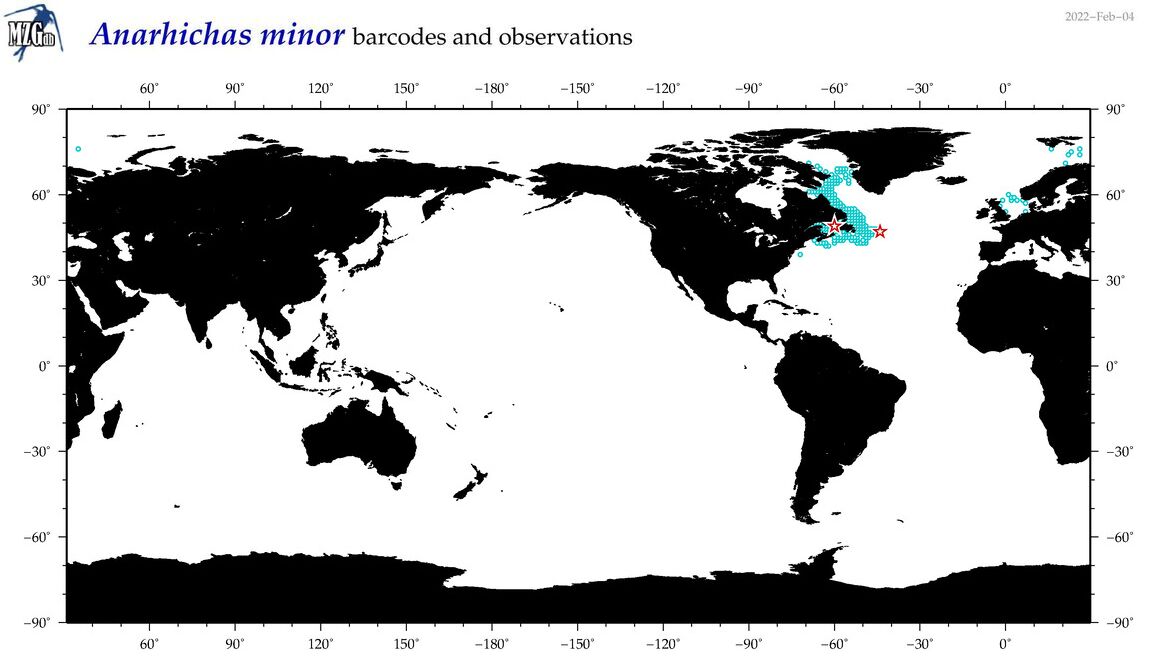 |
accepted
T5003091
R:1:0:0:0 |
| Anisarchus medius |
Species
(7) |
COI
12S
16S
18S
28S
|
COI = 38
12S = 4
16S = 3
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
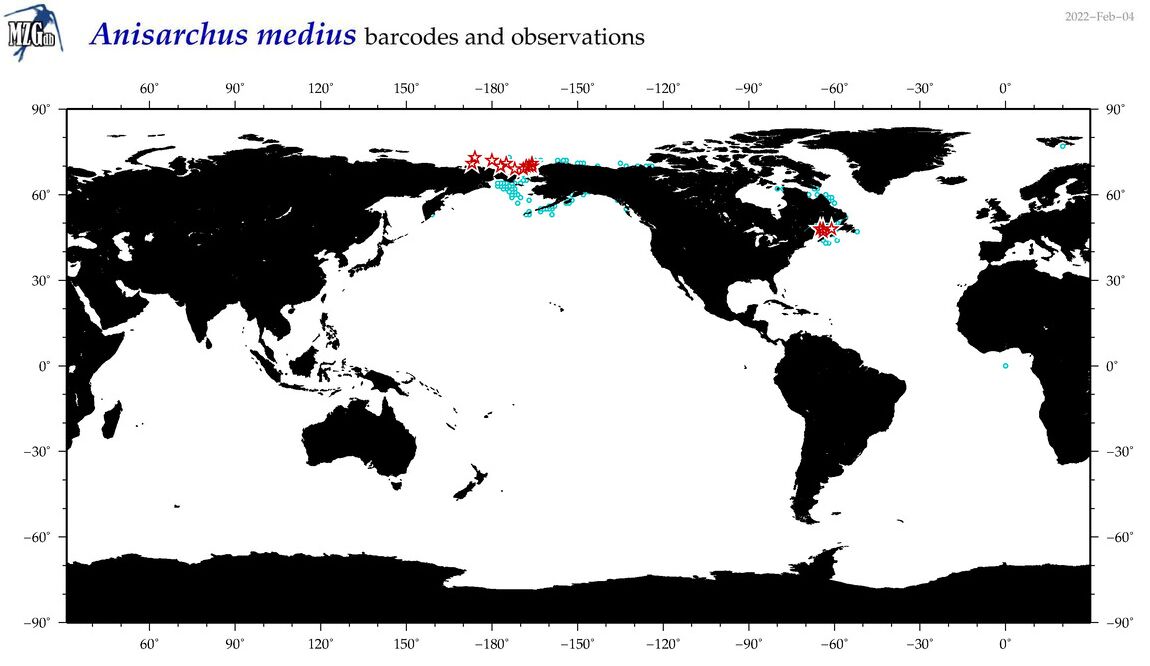 |
accepted
T5000834
R:1:0:0:0 |
| Artediellus atlanticus |
Species
(8) |
COI
12S
16S
18S
28S
|
COI = 31
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
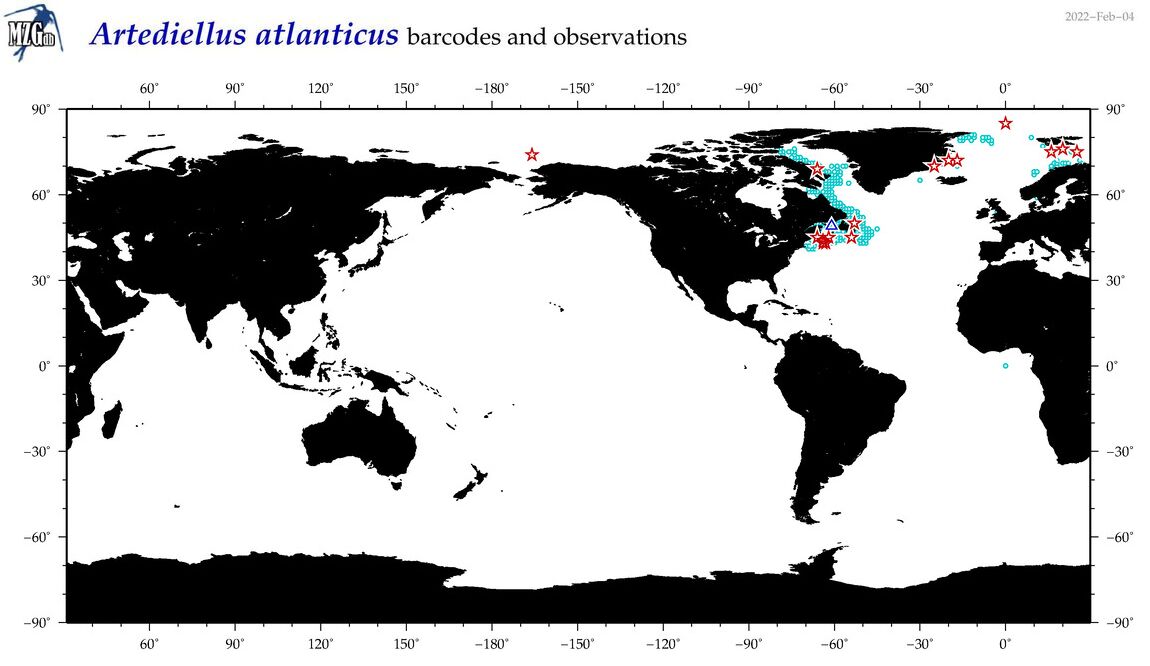 |
accepted
T5003186
R:1:0:0:0 |
| Careproctus longipinnis |
Species
(9) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
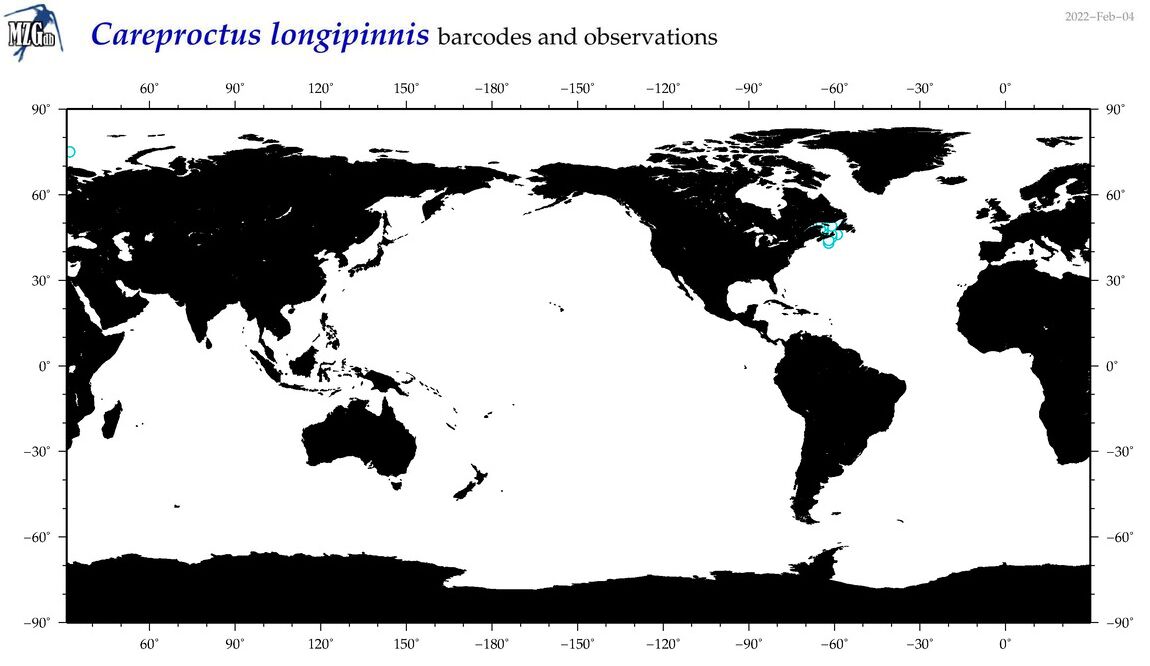 |
accepted
T5017619
R:1:0:0:0 |
| Careproctus reinhardti |
Species
(10) |
COI
12S
16S
18S
28S
|
COI = 8
12S = 2
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
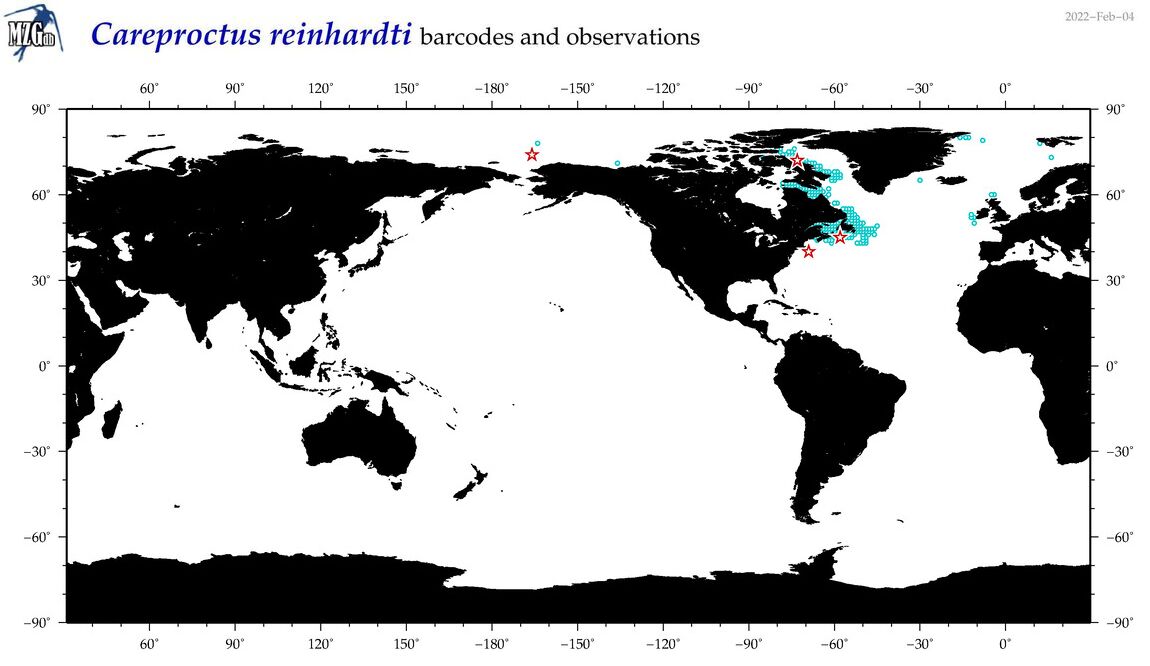 |
accepted
T5003408
R:1:0:0:0 |
| Chirolophis ascanii |
Species
(11) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 2
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
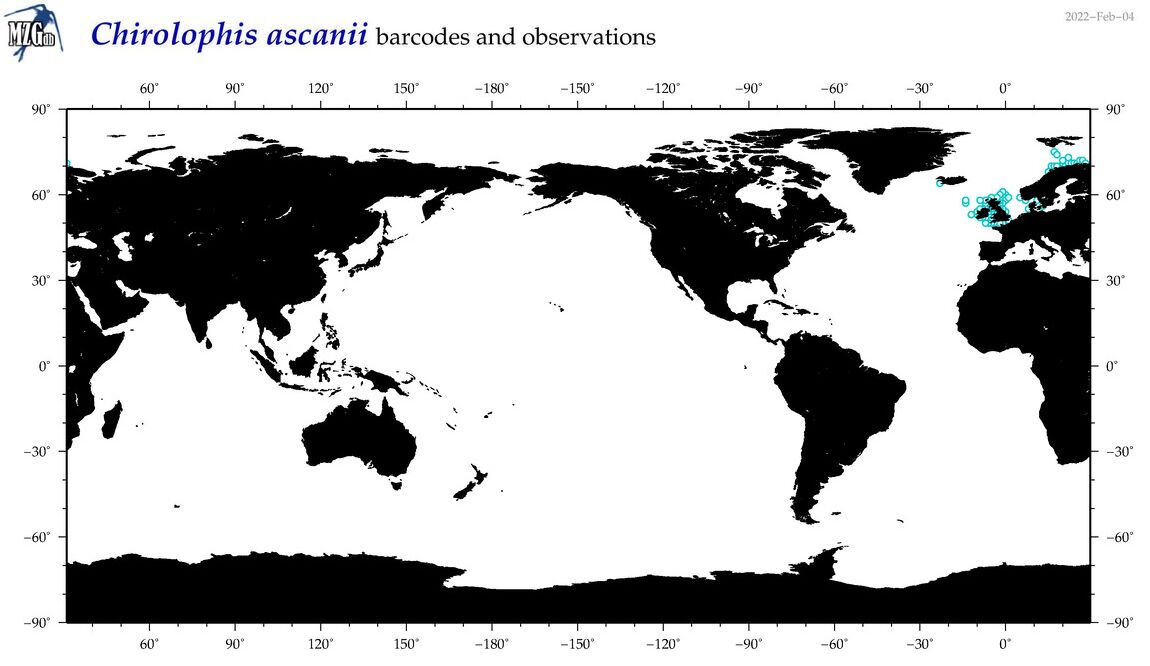 |
accepted
T5018037
R:1:0:0:0 |
| Cottunculus microps |
Species
(12) |
COI
12S
16S
18S
28S
|
COI = 10
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
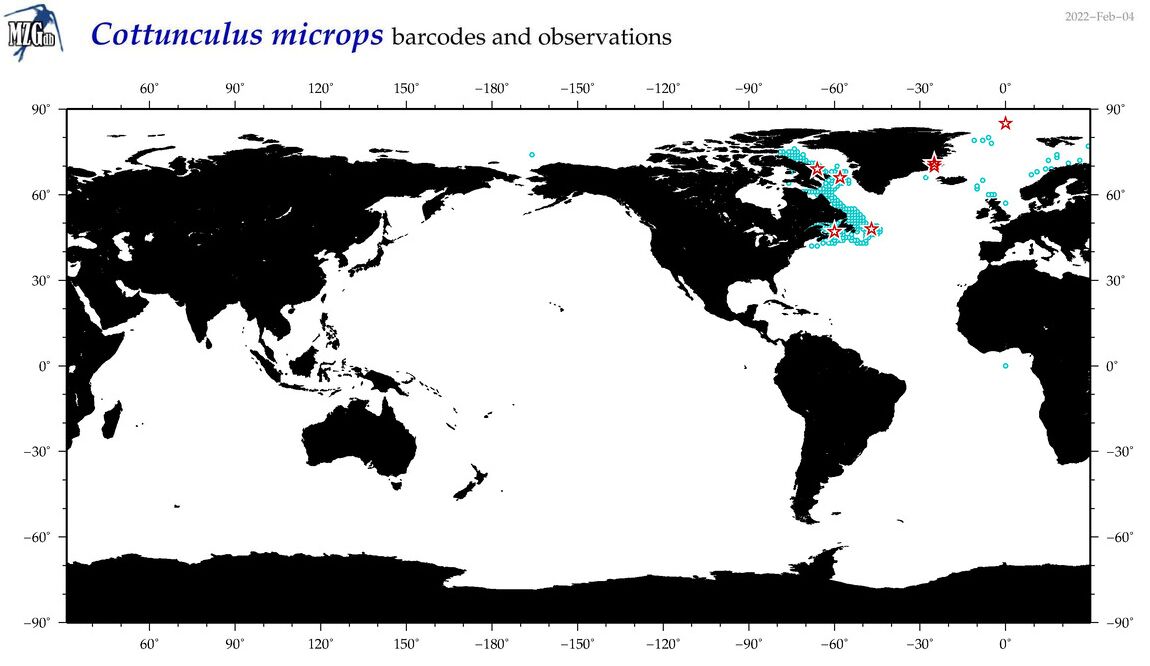 |
accepted
T5003656
R:1:0:0:0 |
| Cyclopterus lumpus |
Species
(13) |
COI
12S
16S
18S
28S
|
COI = 67
12S = 6
16S = 9
18S = 0
28S = 2
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
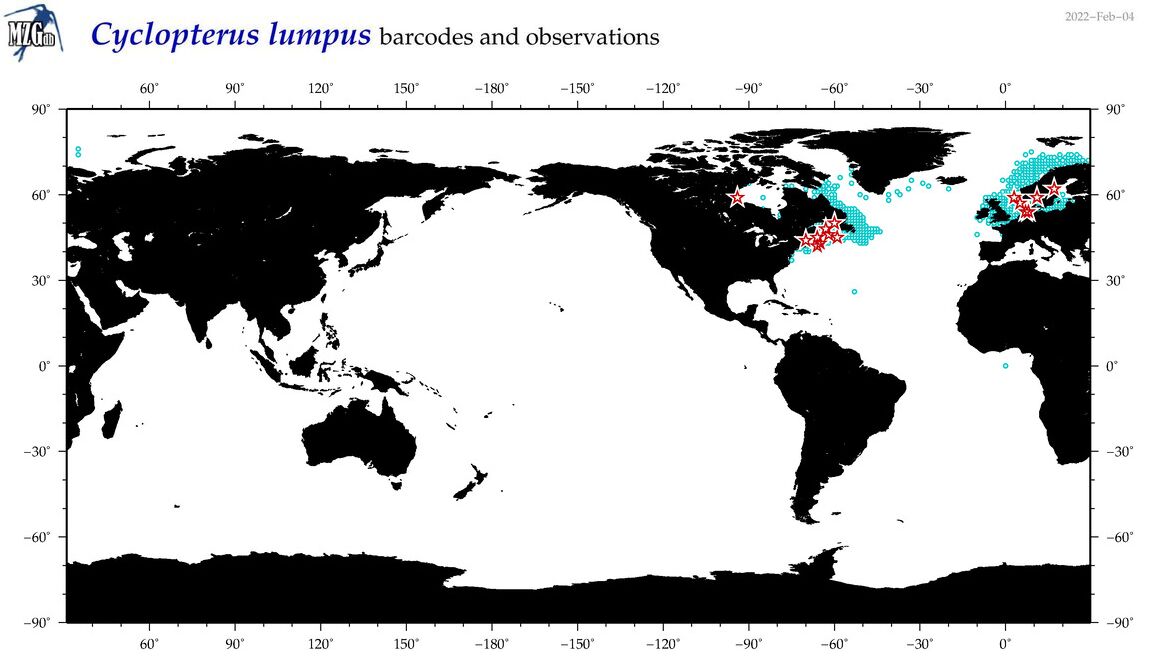 |
accepted
T5000452
R:1:0:0:0 |
| Eumicrotremus derjugini |
Species
(14) |
COI
12S
16S
18S
28S
|
COI = 11
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
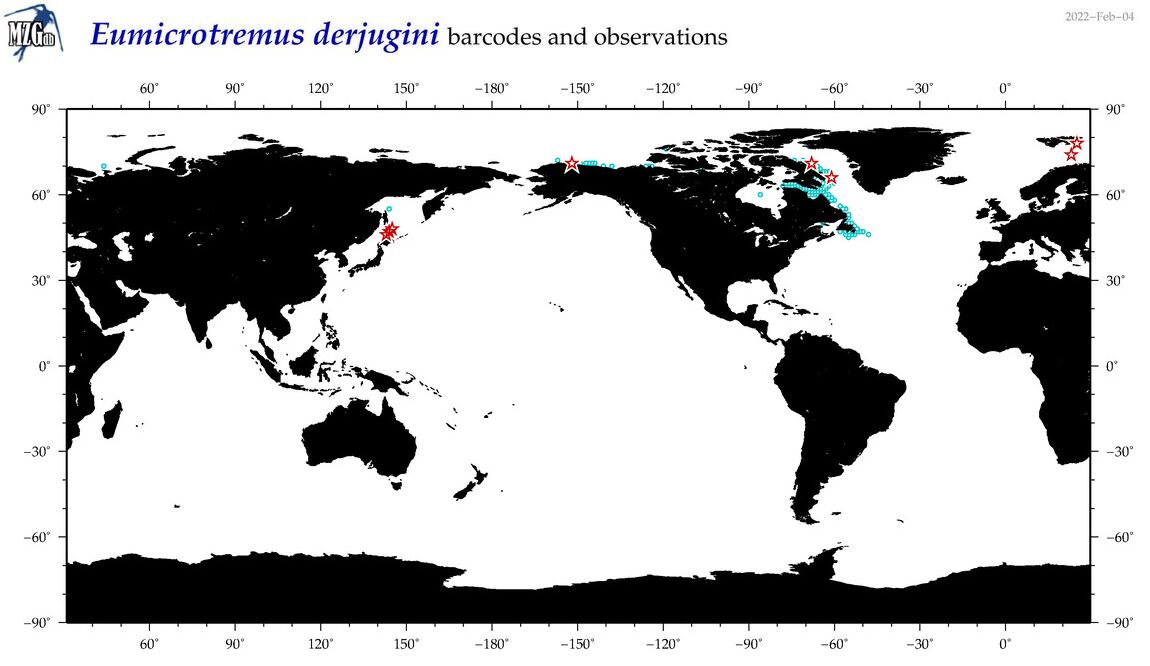 |
accepted
T5003976
R:1:0:0:0 |
| Eumicrotremus spinosus |
Species
(15) |
COI
12S
16S
18S
28S
|
COI = 23
12S = 5
16S = 3
18S = 0
28S = 0
|
COI = 8
12S = 0
16S = 0
18S = 0
28S = 0
|
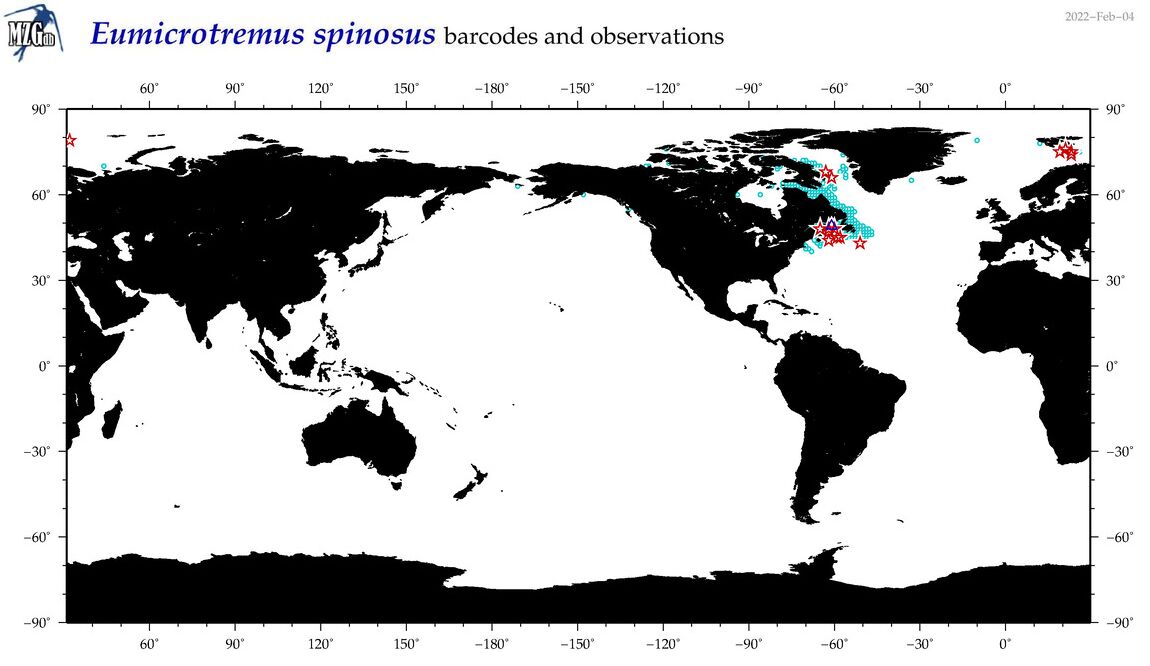 |
accepted
T5003983
R:1:0:0:0 |
| Eutrigla gurnardus |
Species
(16) |
COI
12S
16S
18S
28S
|
COI = 28
12S = 1
16S = 6
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
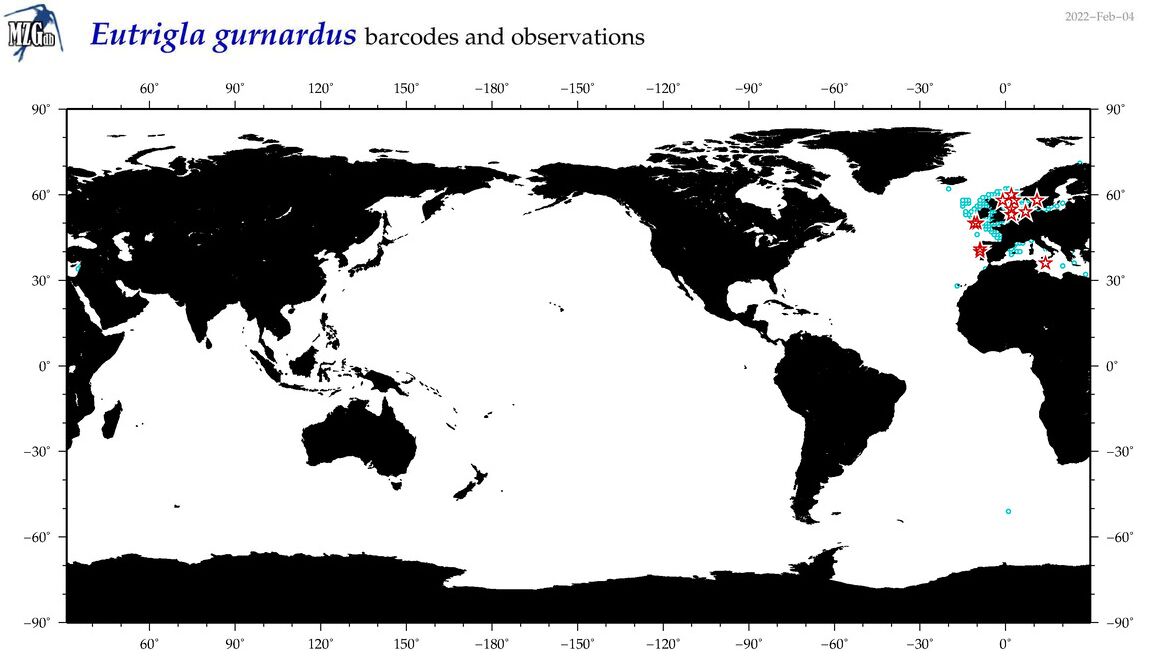 |
accepted
T5003990
R:1:1:0:0 |
| Gasterosteus aculeatus |
Species
(17) |
COI
12S
16S
18S
28S
|
COI = 355
12S = 27
16S = 29
18S = 0
28S = 1
|
COI = 15
12S = 0
16S = 0
18S = 0
28S = 0
|
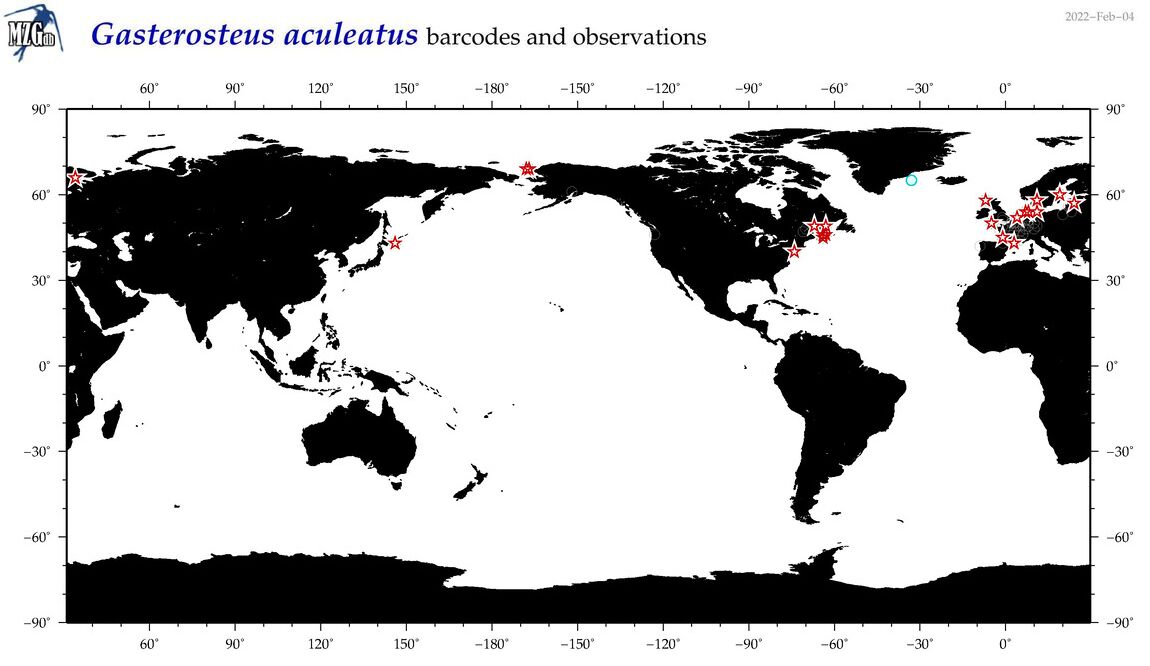 |
accepted
T5000028
R:1:1:1:0 |
| Gymnelus andersoni |
Species
(18) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
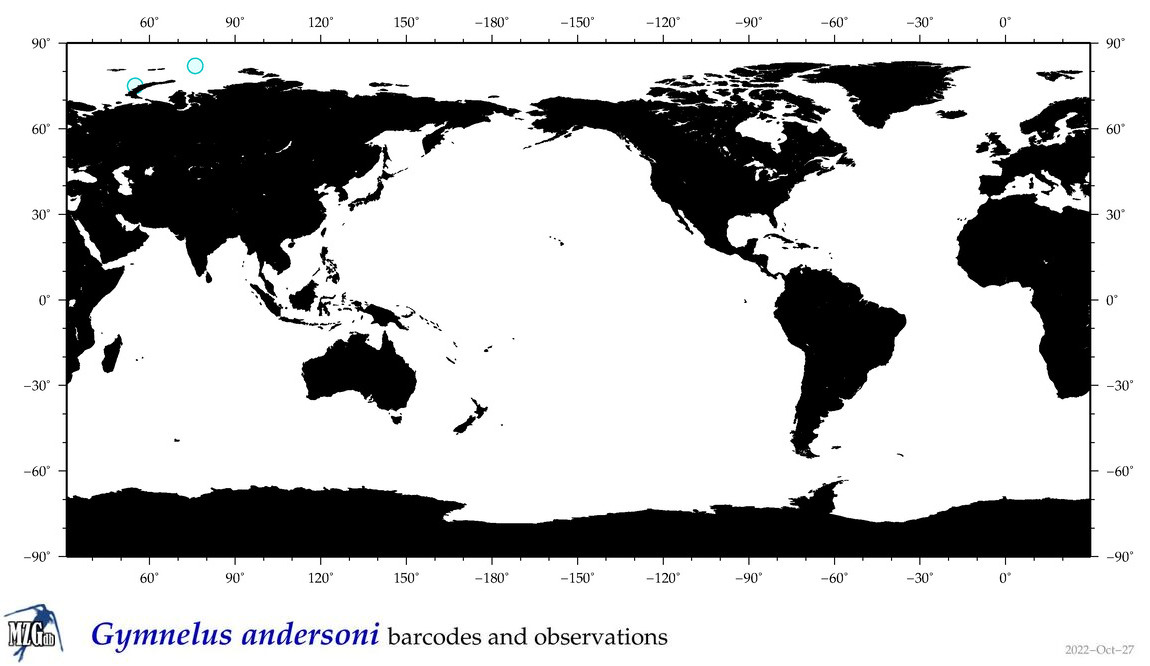 |
accepted
T5028060
R:1:0:0:0 |
| Gymnelus hemifasciatus |
Species
(19) |
COI
12S
16S
18S
28S
|
COI = 48
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004100
R:1:0:0:0 |
| Gymnocanthus pistilliger |
Species
(20) |
COI
12S
16S
18S
28S
|
COI = 26
12S = 11
16S = 10
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
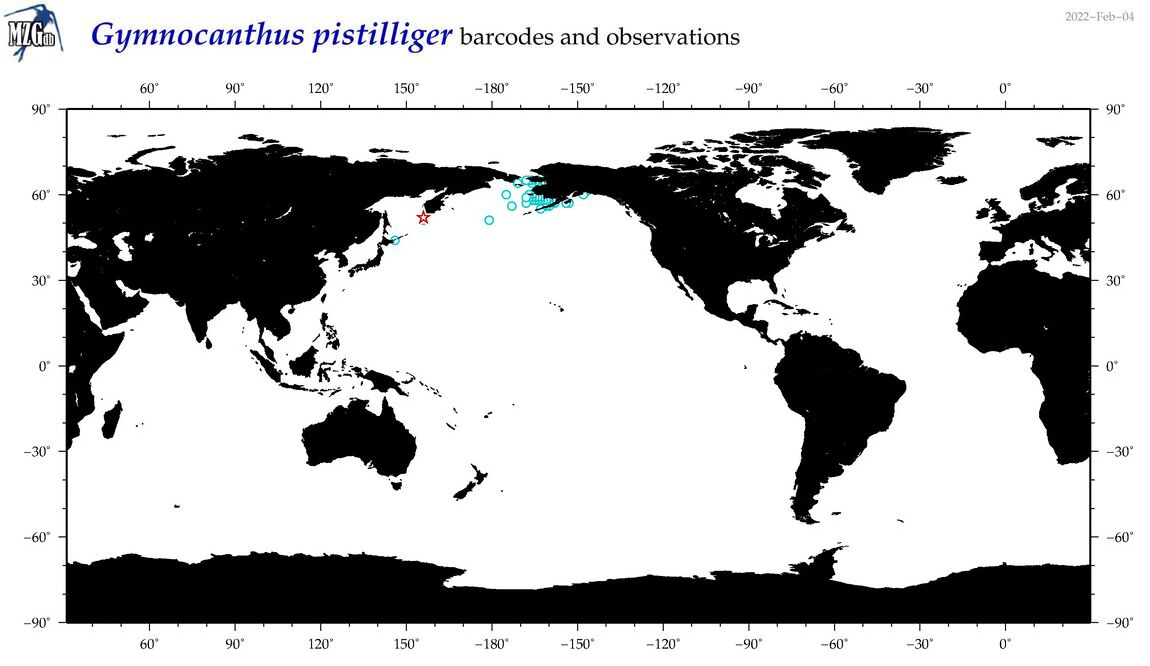 |
accepted
T5004106
R:1:0:0:0 |
| Icelus bicornis |
Species
(21) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
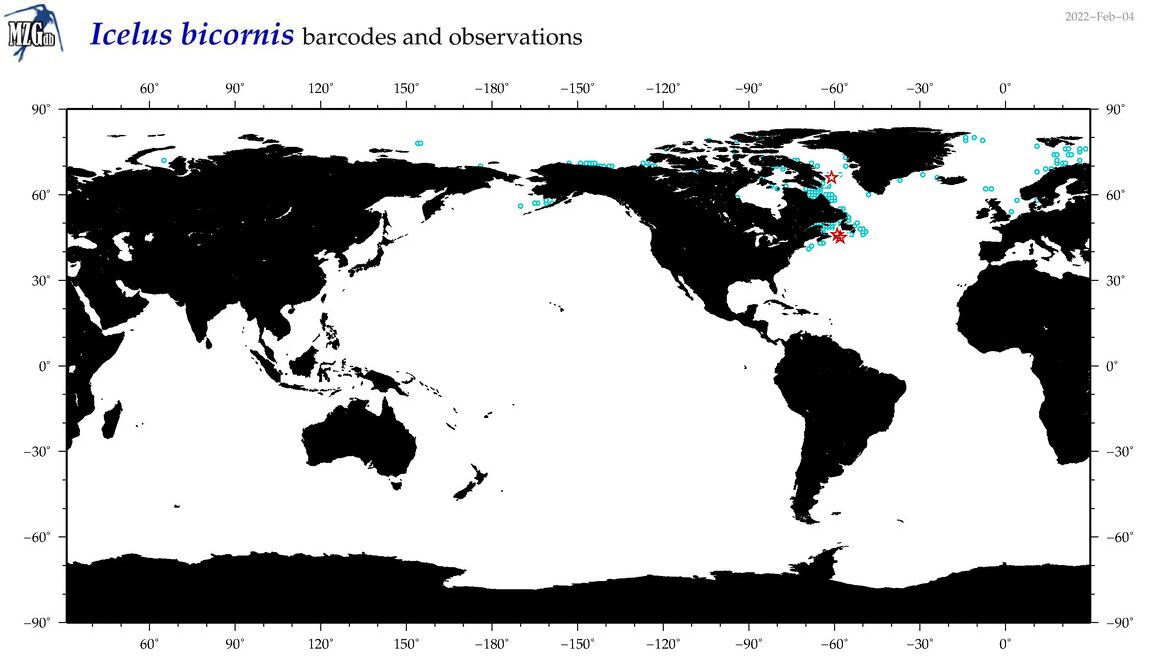 |
accepted
T5004342
R:1:0:0:0 |
| Leptagonus decagonus |
Species
(22) |
COI
12S
16S
18S
28S
|
COI = 21
12S = 3
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
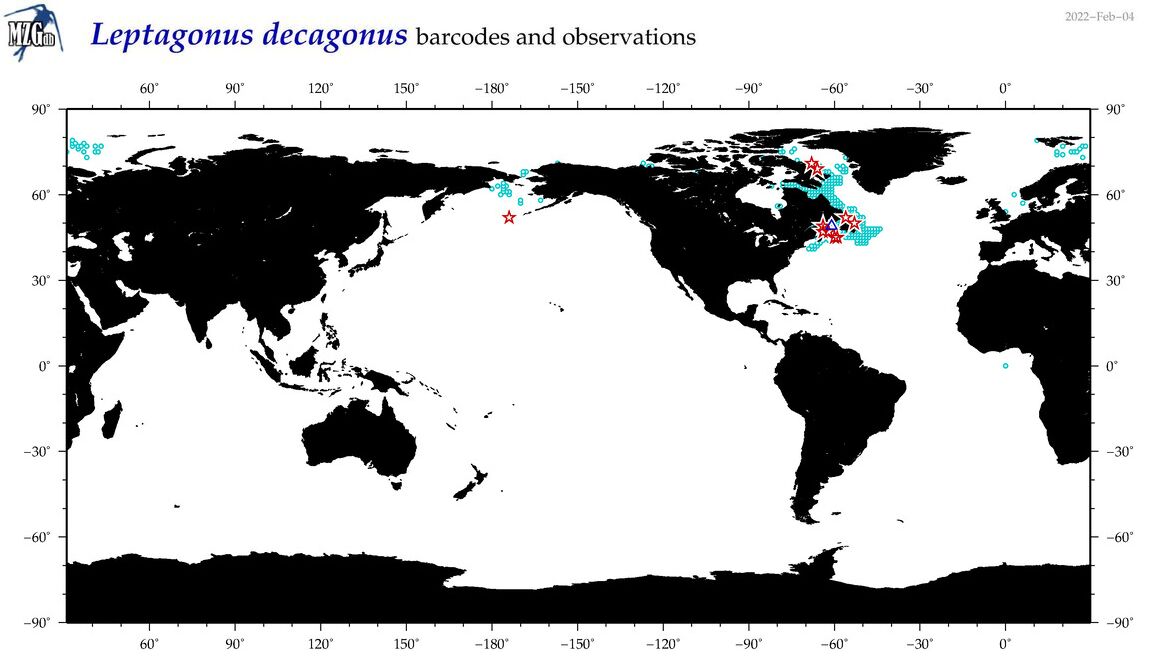 |
accepted
T5004464
R:1:0:0:0 |
| Leptoclinus maculatus |
Species
(23) |
COI
12S
16S
18S
28S
|
COI = 46
12S = 8
16S = 4
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
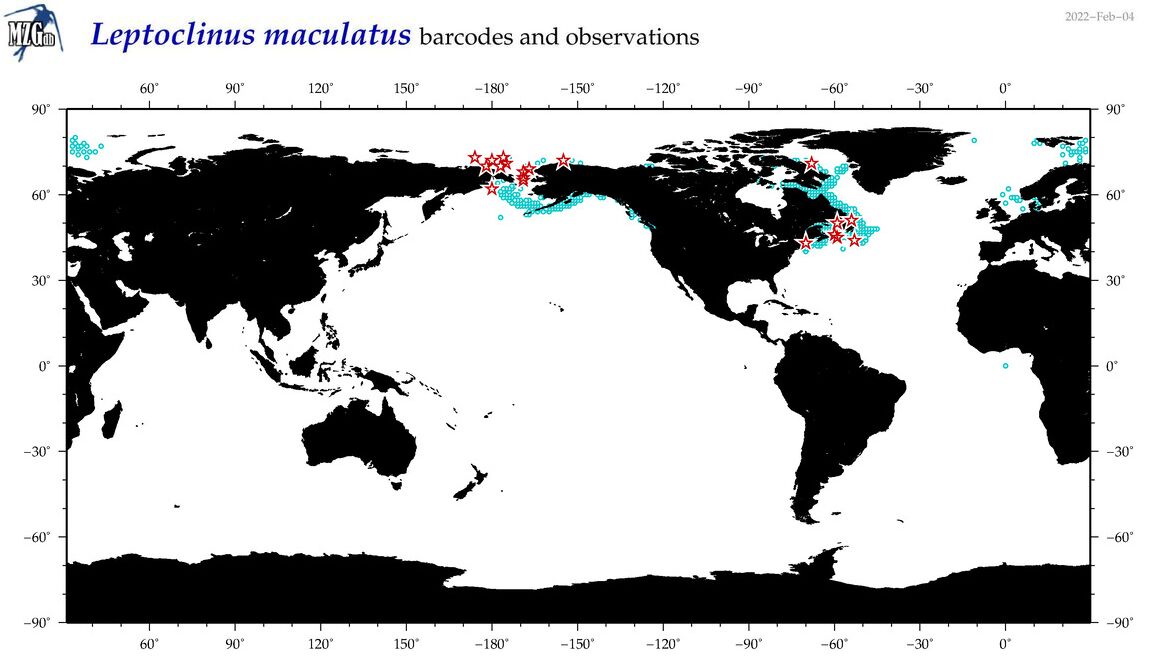 |
accepted
T5000833
R:1:0:0:0 |
| Liparis agassizii |
Species
(24) |
COI
12S
16S
18S
28S
|
COI = 14
12S = 4
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
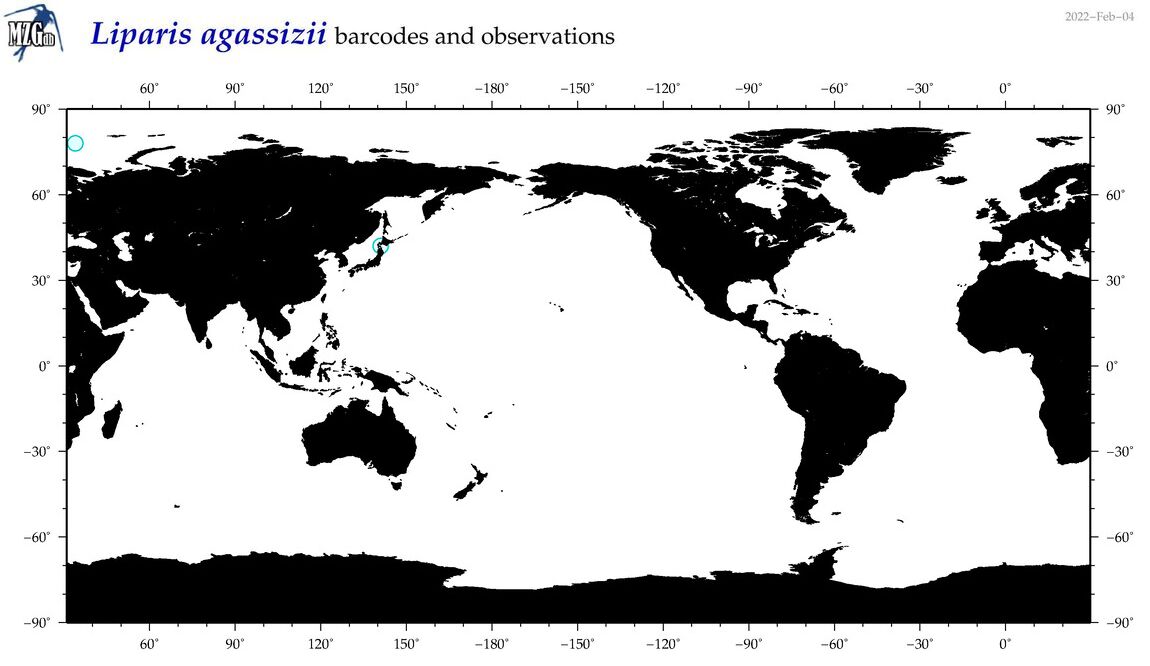 |
accepted
T5004498
R:1:0:0:0 |
| Liparis coheni |
Species
(25) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000806
R:1:0:0:0 |
| Liparis fabricii |
Species
(26) |
COI
12S
16S
18S
28S
|
COI = 23
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
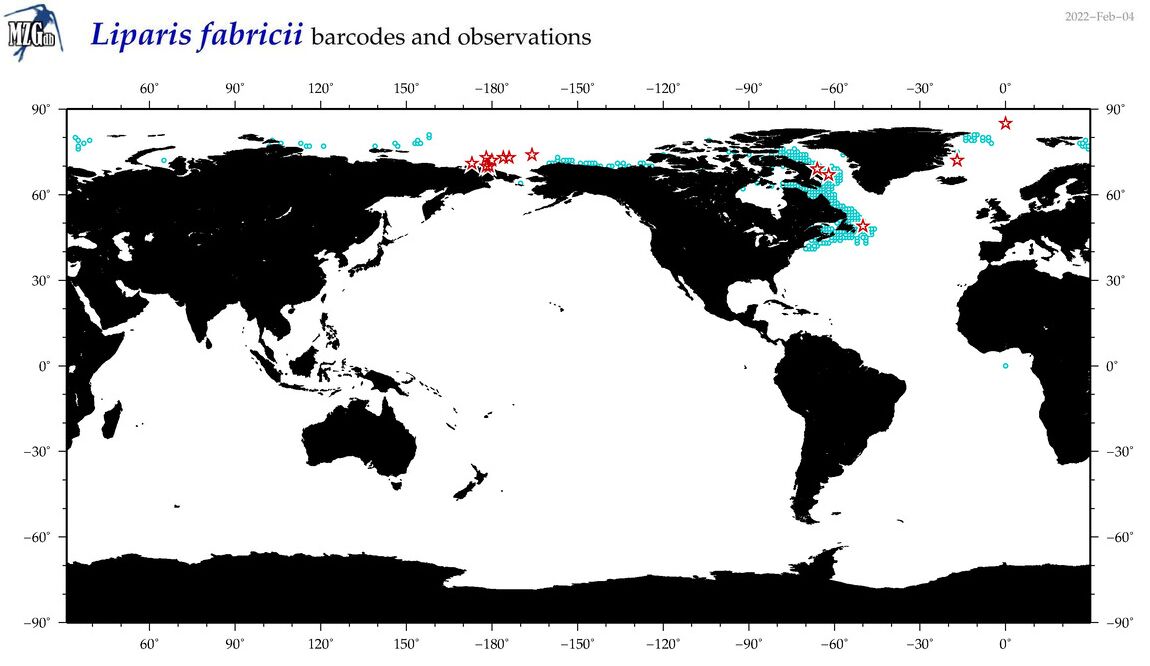 |
accepted
T5000807
R:1:0:0:0 |
| Liparis gibbus |
Species
(27) |
COI
12S
16S
18S
28S
|
COI = 29
12S = 4
16S = 2
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
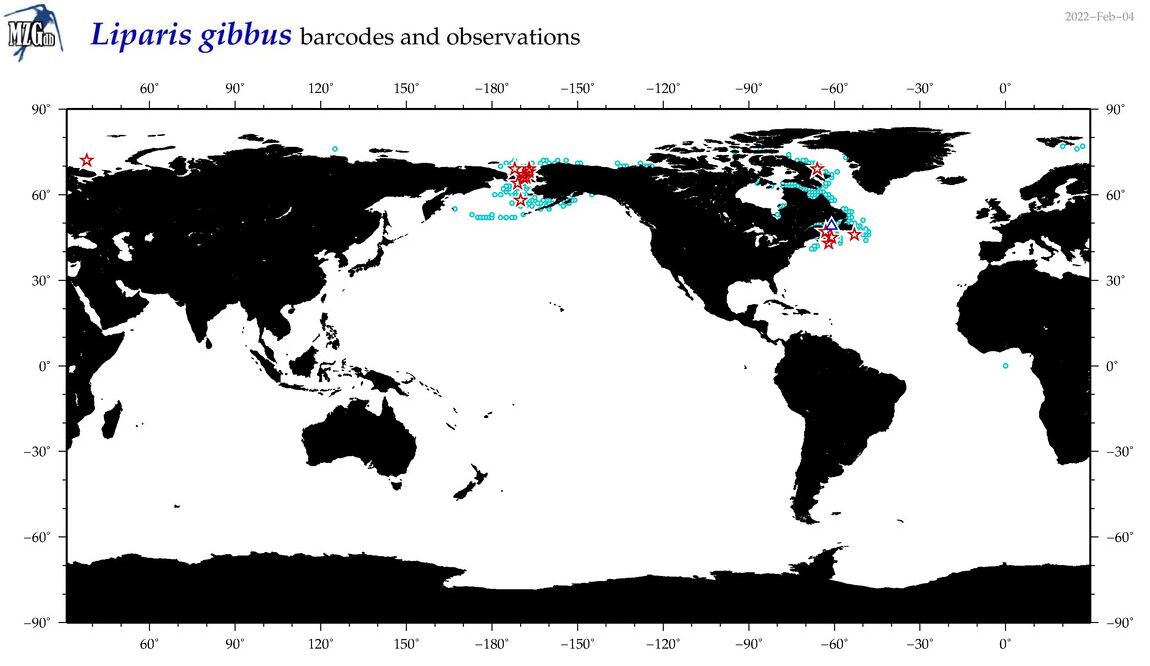 |
accepted
T5000810
R:1:0:0:0 |
| Liparis liparis |
Species
(28) |
COI
12S
16S
18S
28S
|
COI = 15
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004505
R:1:1:0:0 |
| Liparis montagui |
Species
(29) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
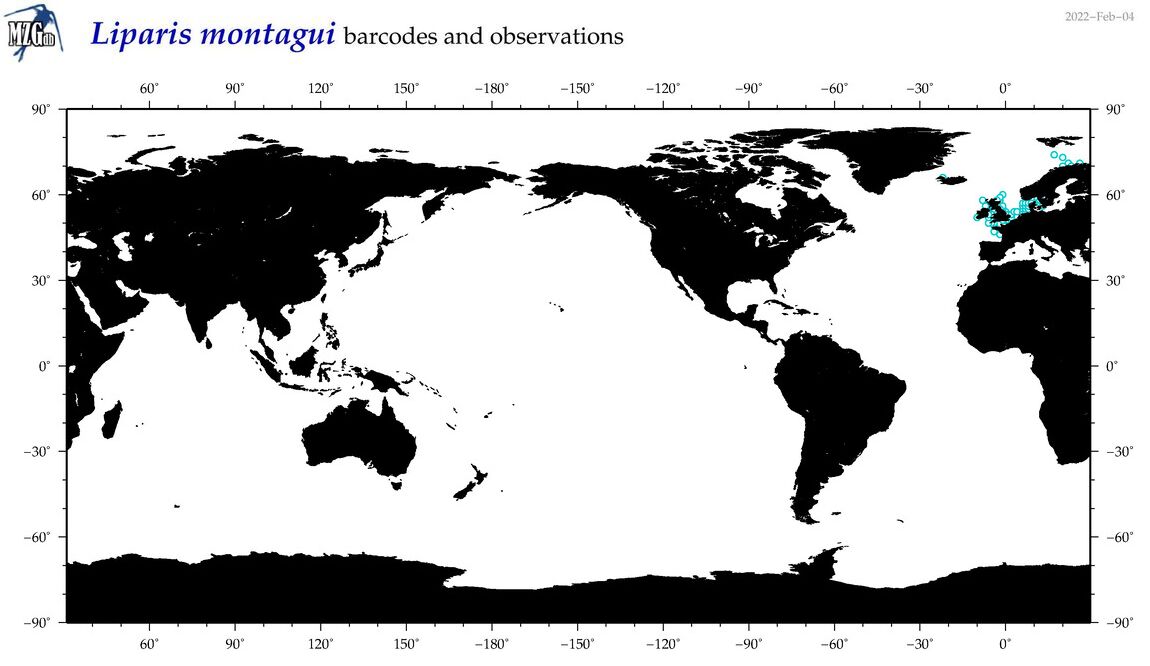 |
accepted
T5004506
R:1:0:0:0 |
| Lumpenus lampretaeformis |
Species
(30) |
COI
12S
16S
18S
28S
|
COI = 24
12S = 3
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
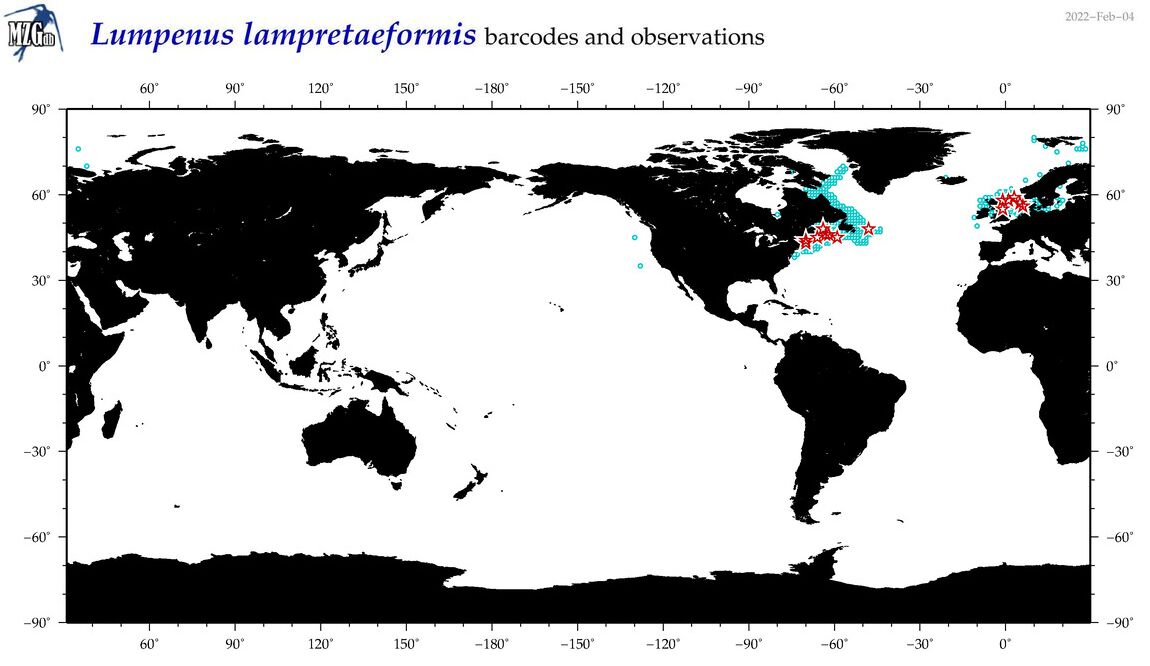 |
accepted
T5000832
R:1:0:0:0 |
| Lycenchelys muraena |
Species
(31) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 5
16S = 3
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
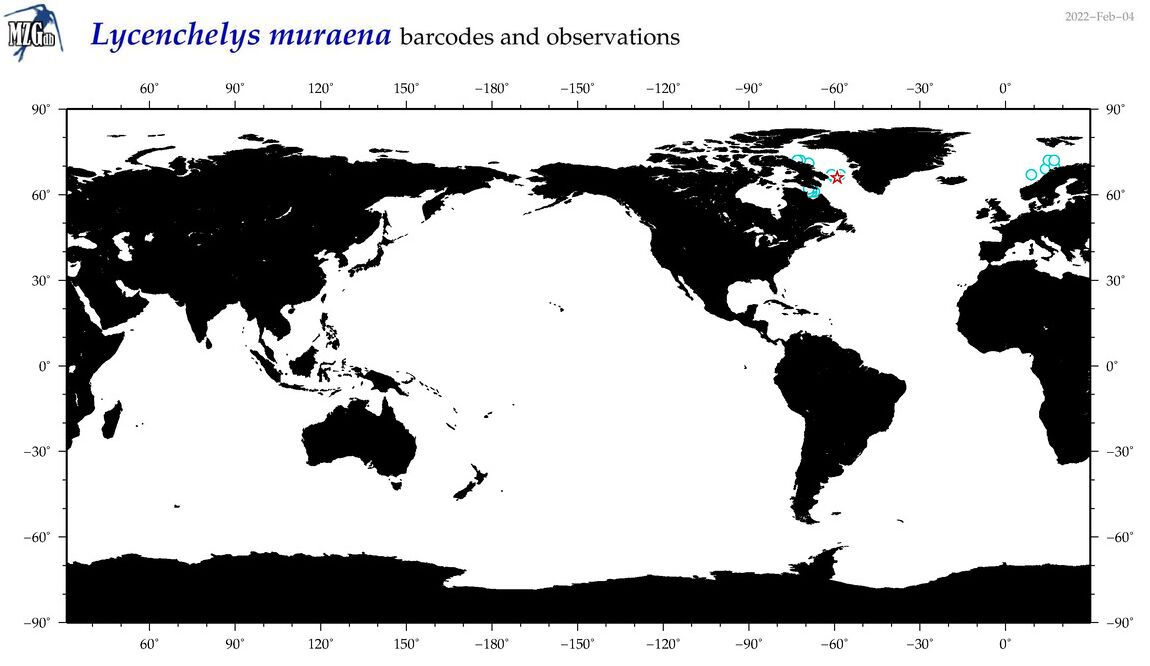 |
accepted
T5004551
R:1:0:0:0 |
| Lycenchelys sarsii |
Species
(32) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 3
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
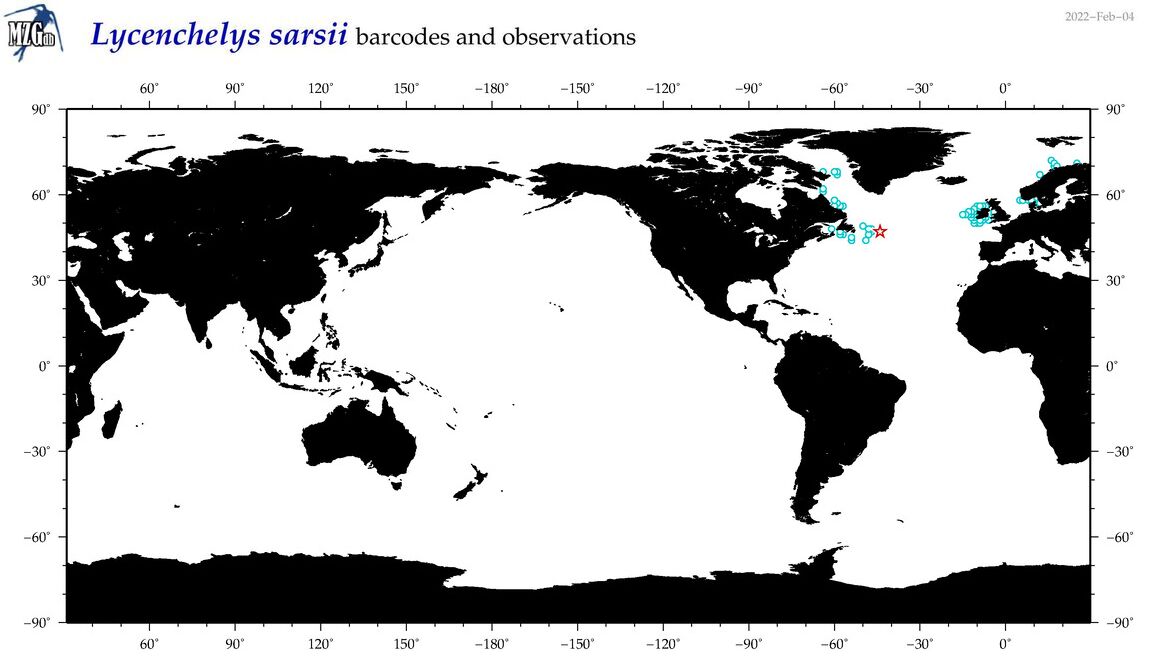 |
accepted
T5004554
R:1:0:0:0 |
| Lycodes esmarkii |
Species
(33) |
COI
12S
16S
18S
28S
|
COI = 6
12S = 3
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
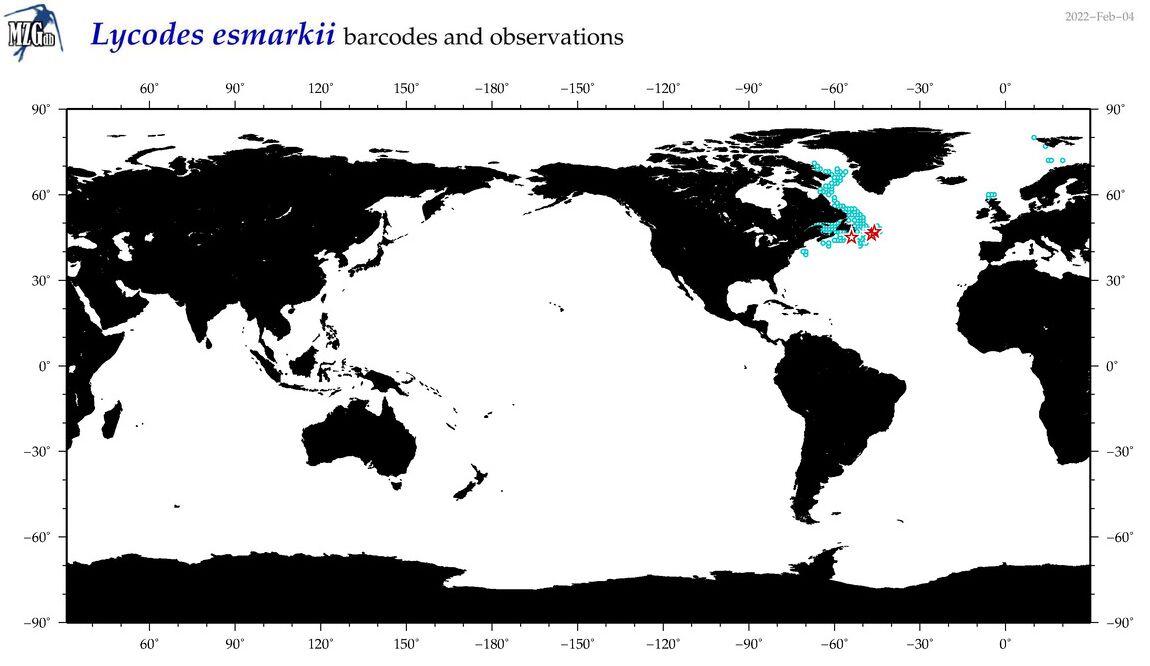 |
accepted
T5004570
R:1:0:0:0 |
| Lycodes eudipleurostictus |
Species
(34) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
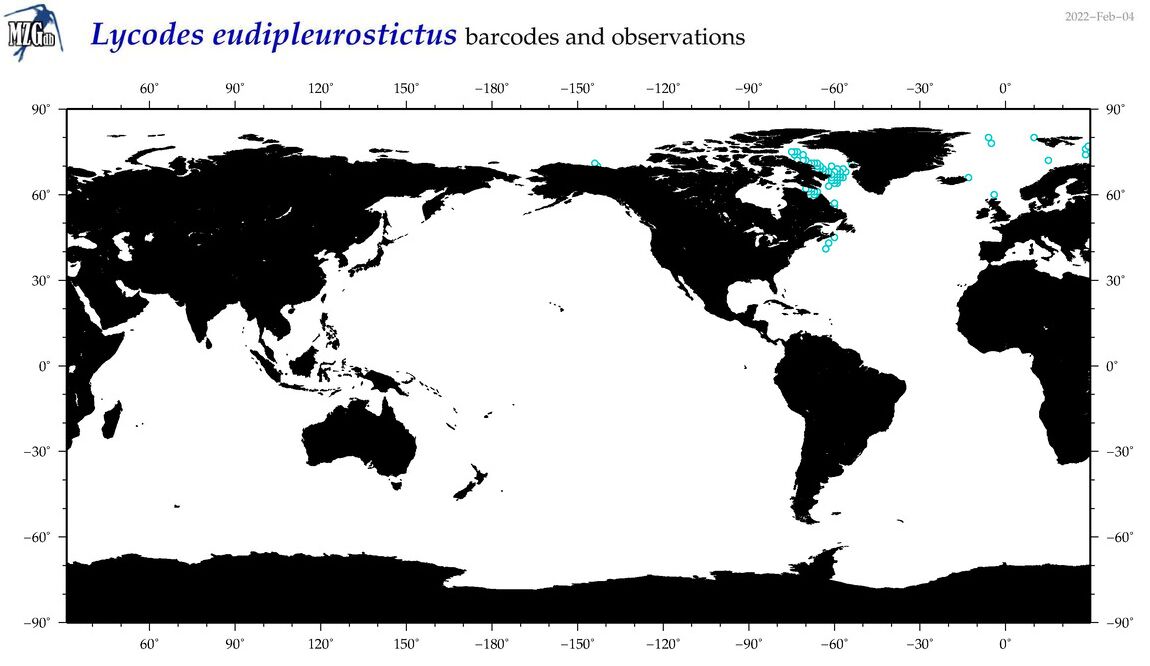 |
accepted
T5021893
R:1:0:0:0 |
| Lycodes gracilis |
Species
(35) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
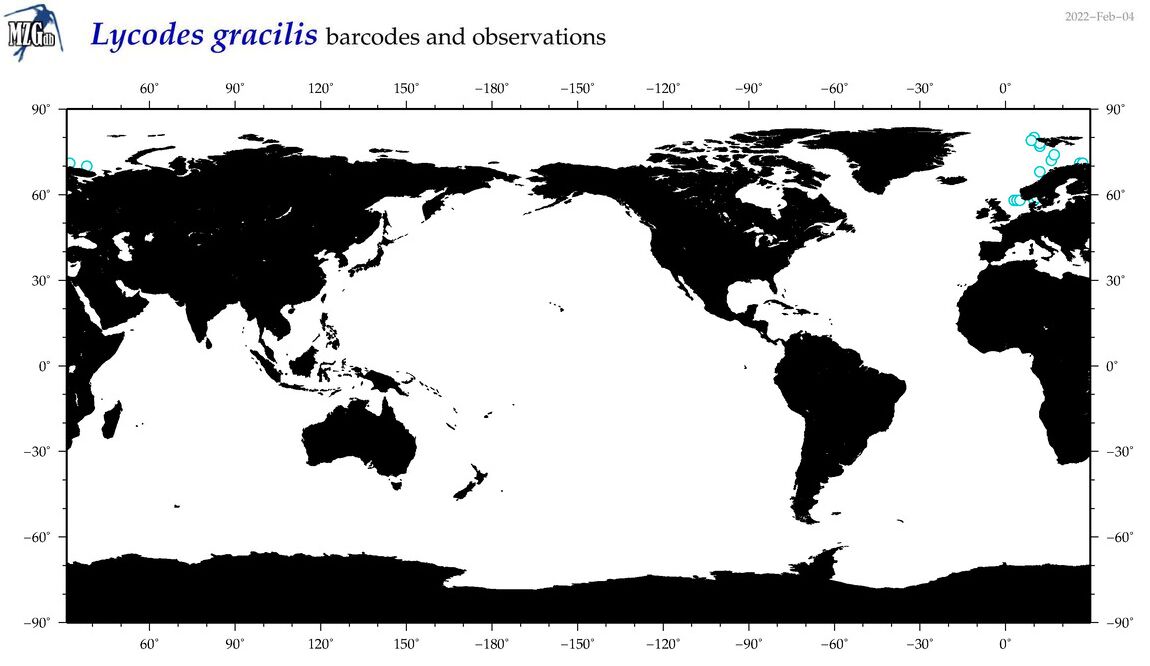 |
accepted
T5021895
R:1:0:0:0 |
| Lycodes luetkenii |
Species
(36) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
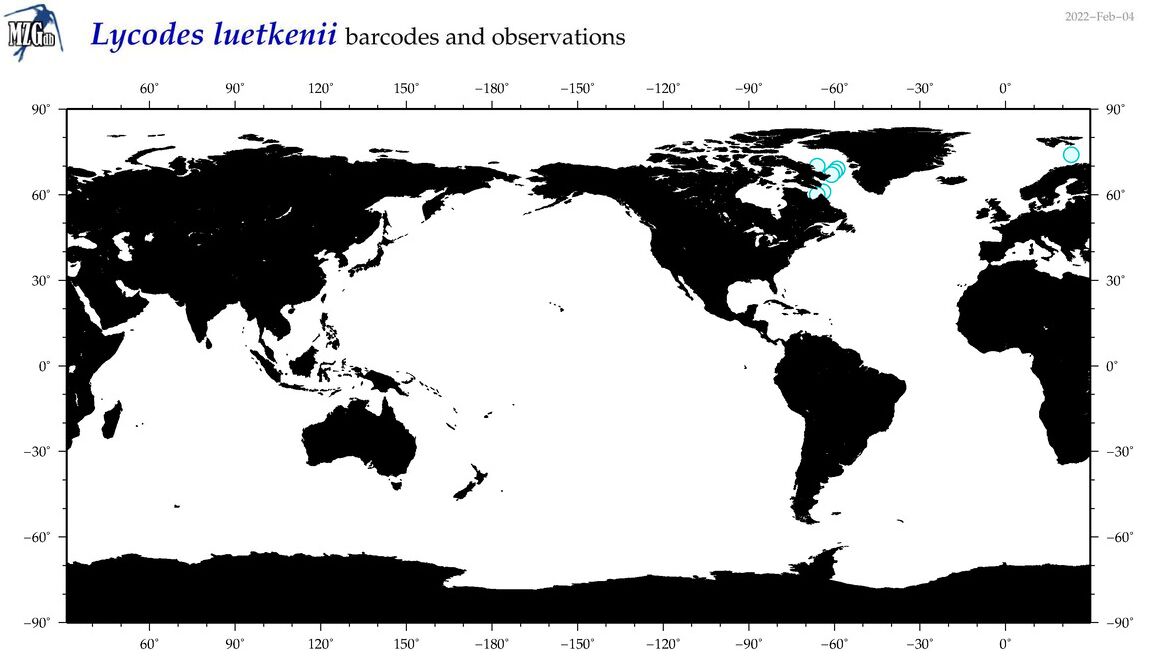 |
accepted
T5021897
R:1:0:0:0 |
| Lycodes paamiuti |
Species
(37) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
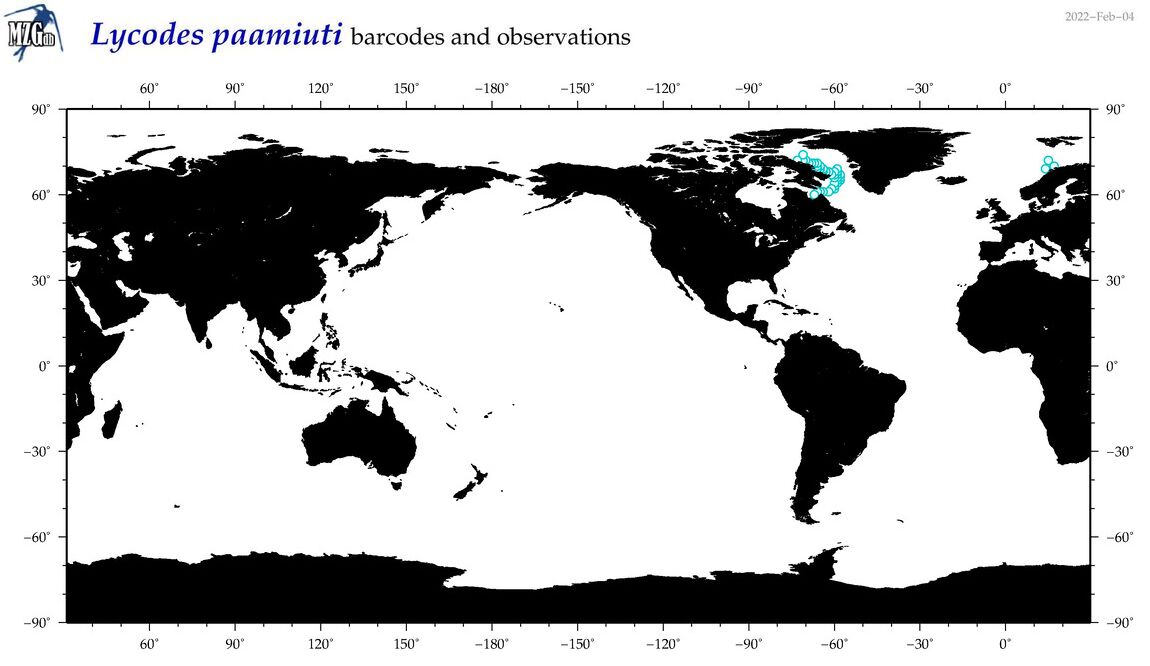 |
accepted
T5021900
R:1:0:0:0 |
| Lycodes pallidus |
Species
(38) |
COI
12S
16S
18S
28S
|
COI = 11
12S = 1
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
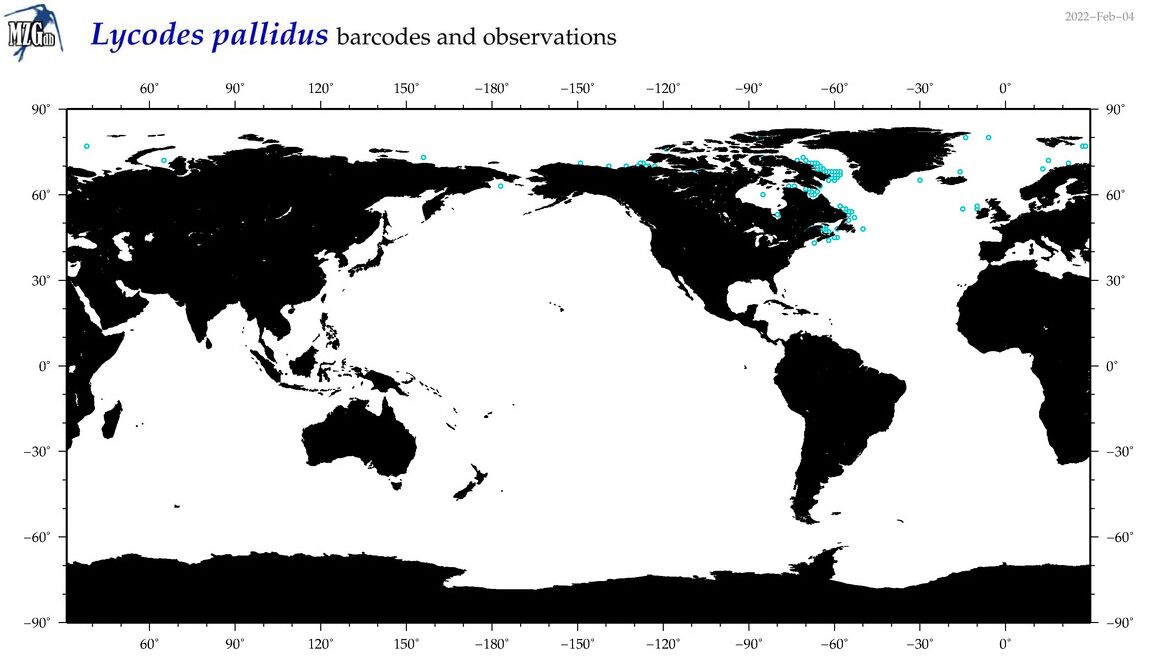 |
accepted
T5004586
R:1:0:0:0 |
| Lycodes reticulatus |
Species
(39) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 5
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
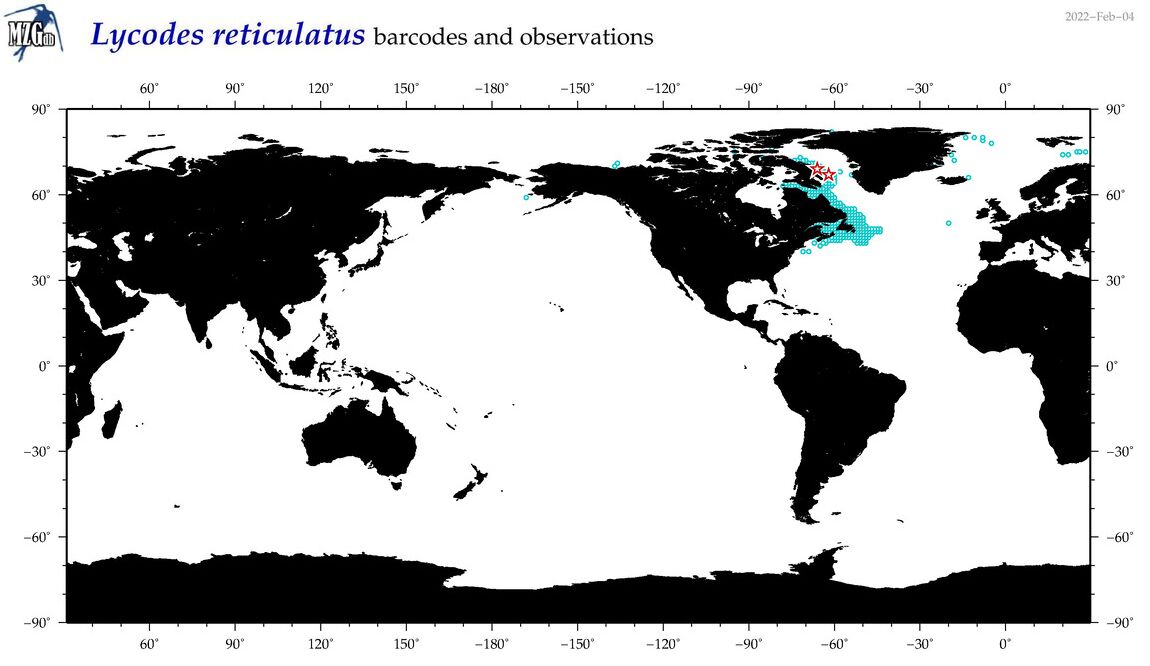 |
accepted
T5004589
R:1:0:0:0 |
| Lycodes rossi |
Species
(40) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 2
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5021903
R:1:0:0:0 |
| Lycodes squamiventer |
Species
(41) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
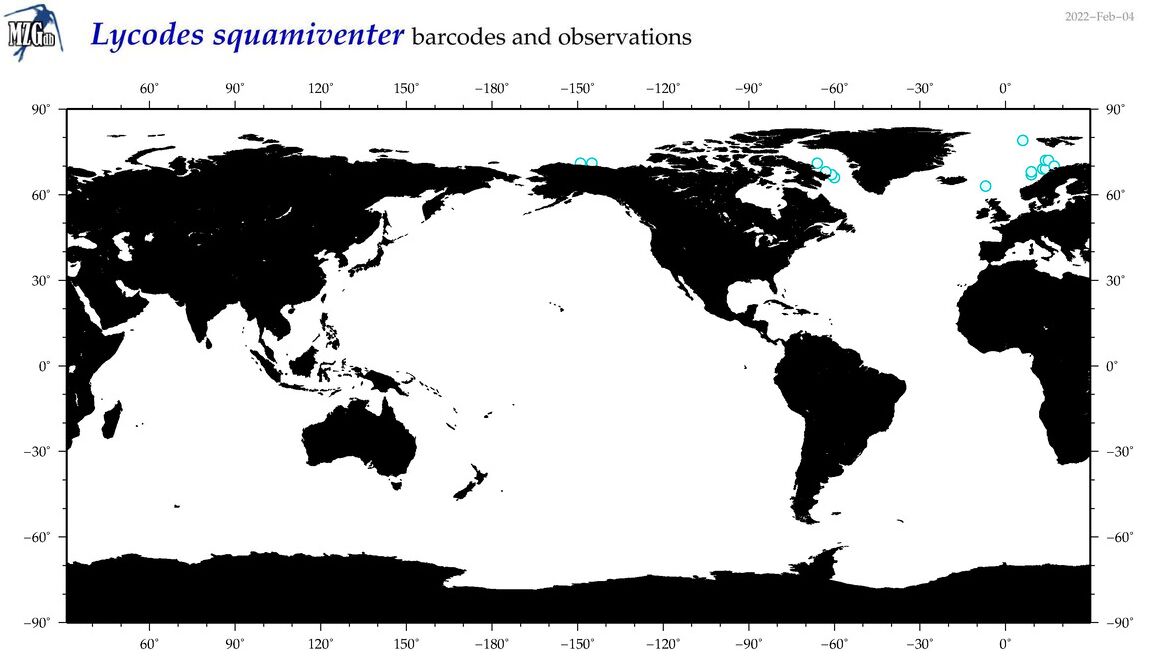 |
accepted
T5021905
R:1:0:0:0 |
| Lycodes vahlii |
Species
(42) |
COI
12S
16S
18S
28S
|
COI = 15
12S = 4
16S = 3
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
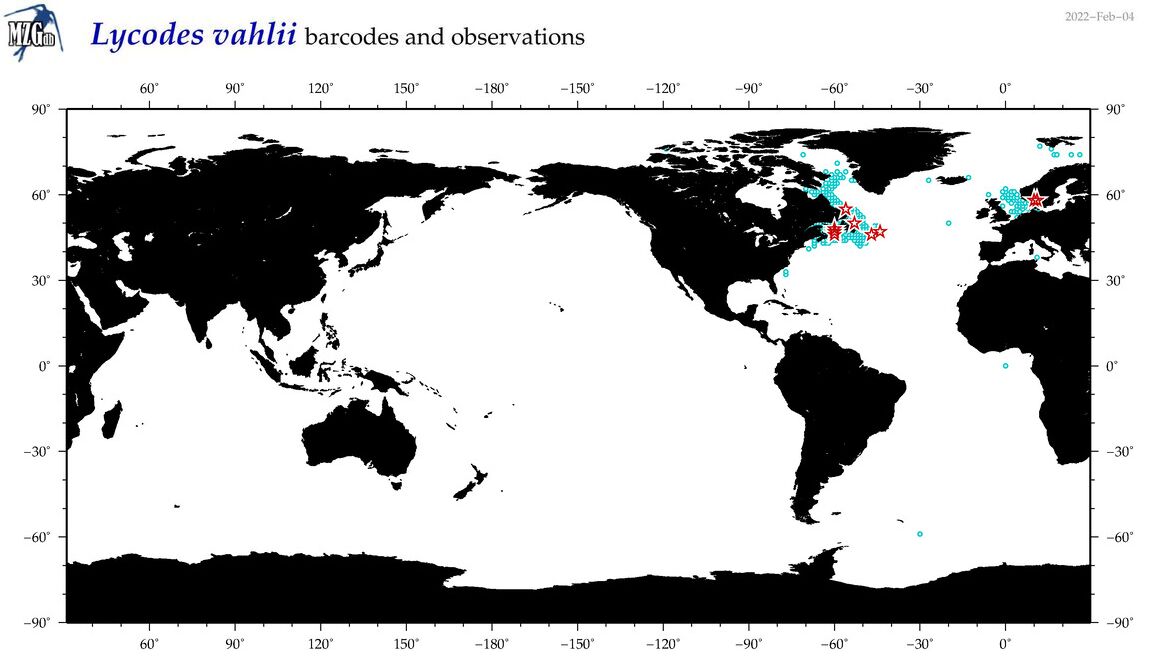 |
accepted
T5004597
R:1:0:0:0 |
| Lycodonus flagellicauda |
Species
(43) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
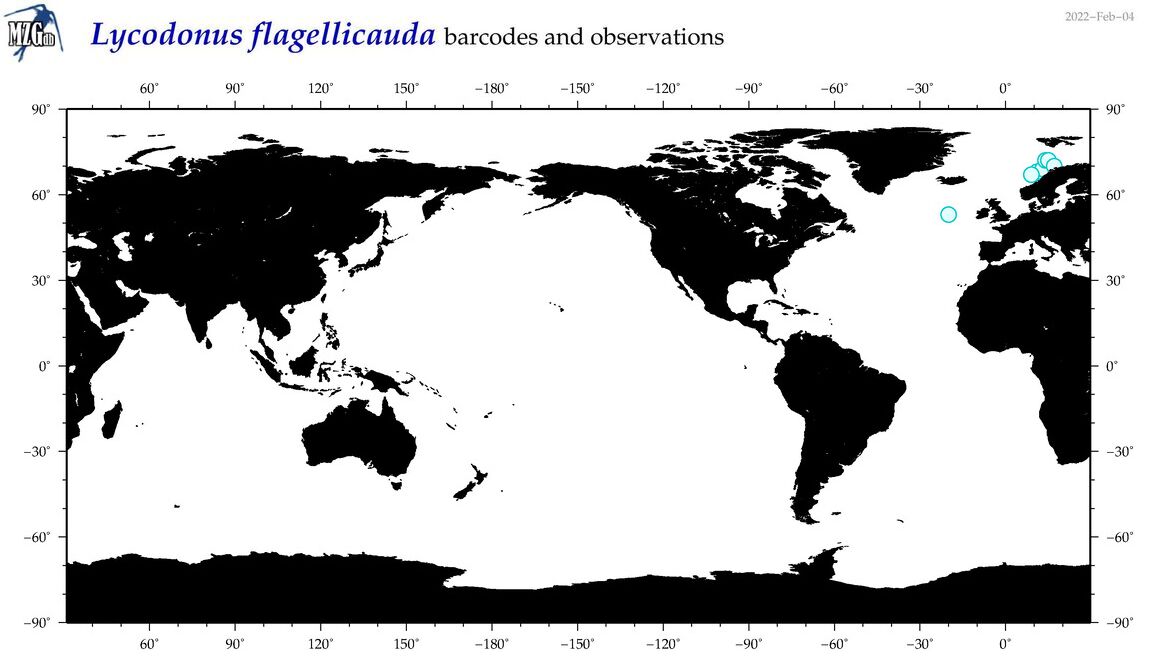 |
accepted
T5021908
R:1:0:0:0 |
| Meganthias natalensis |
Species
(44) |
COI
12S
16S
18S
28S
|
COI = 8
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
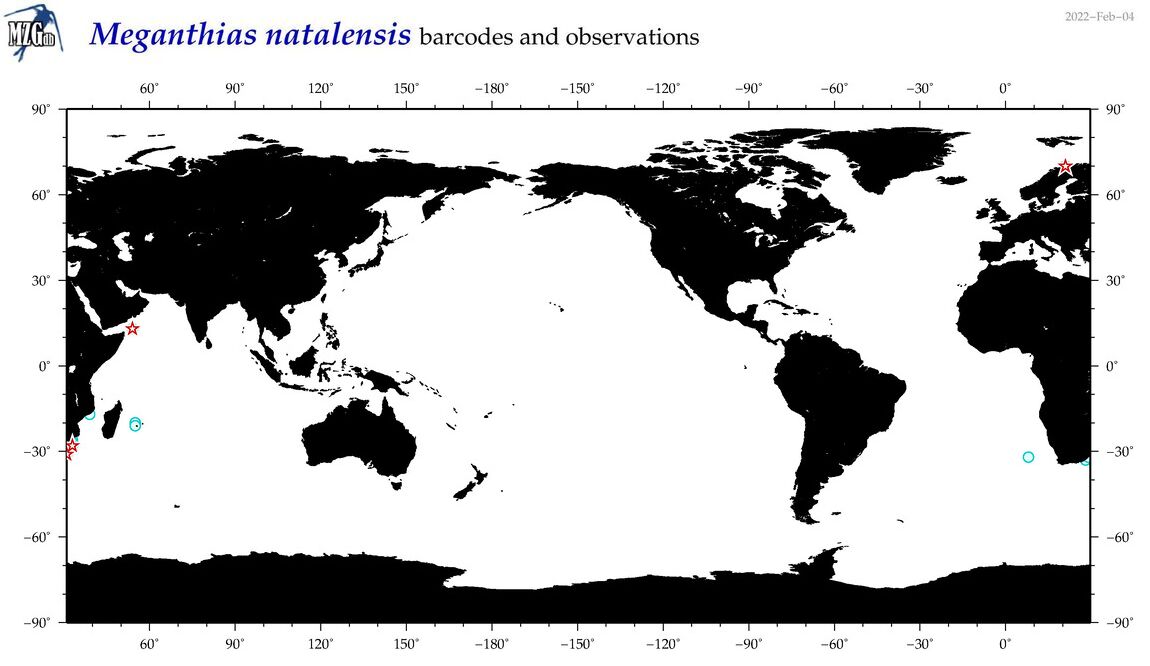 |
accepted
T5004641
R:1:0:0:0 |
| Myoxocephalus quadricornis |
Species
(45) |
COI
12S
16S
18S
28S
|
COI = 45
12S = 8
16S = 7
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
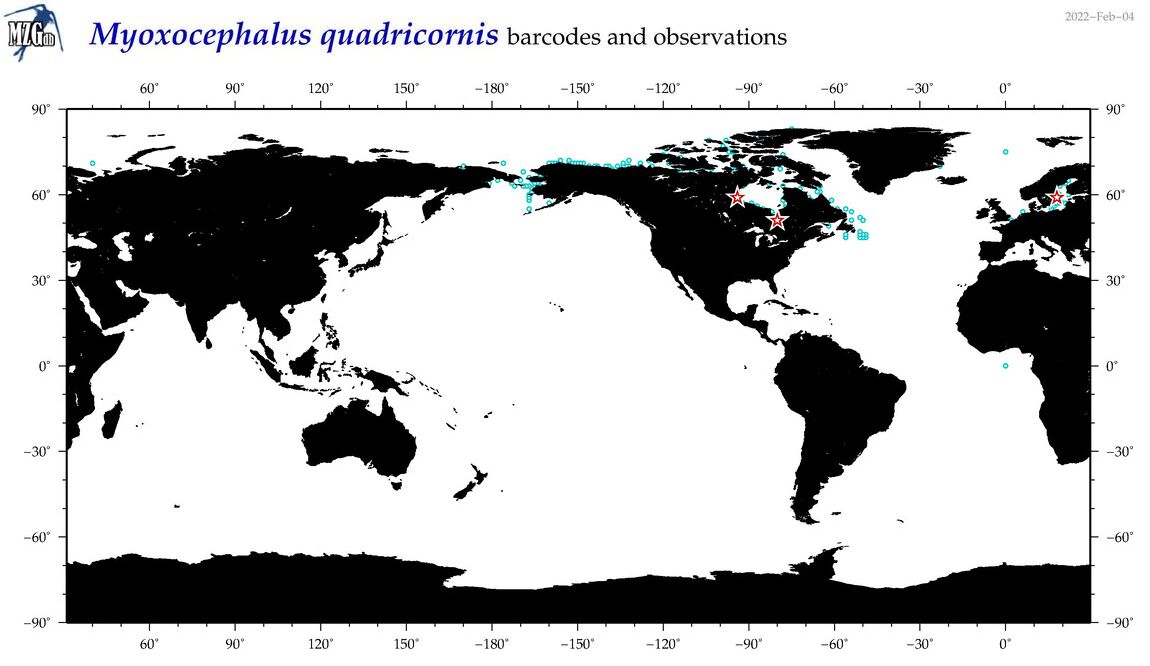 |
accepted
T5004752
R:1:1:1:0 |
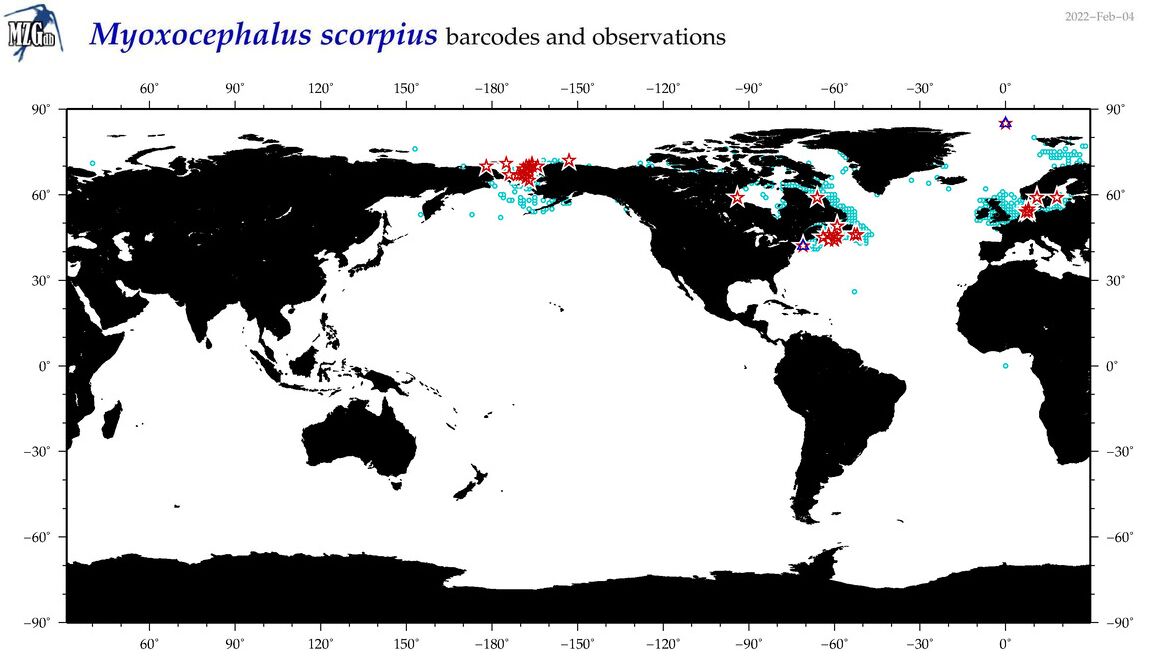
| Myoxocephalus scorpius |
Species
(46) |
COI
12S
16S
18S
28S
|
COI = 85
12S = 6
16S = 6
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000952
R:1:1:0:0 |
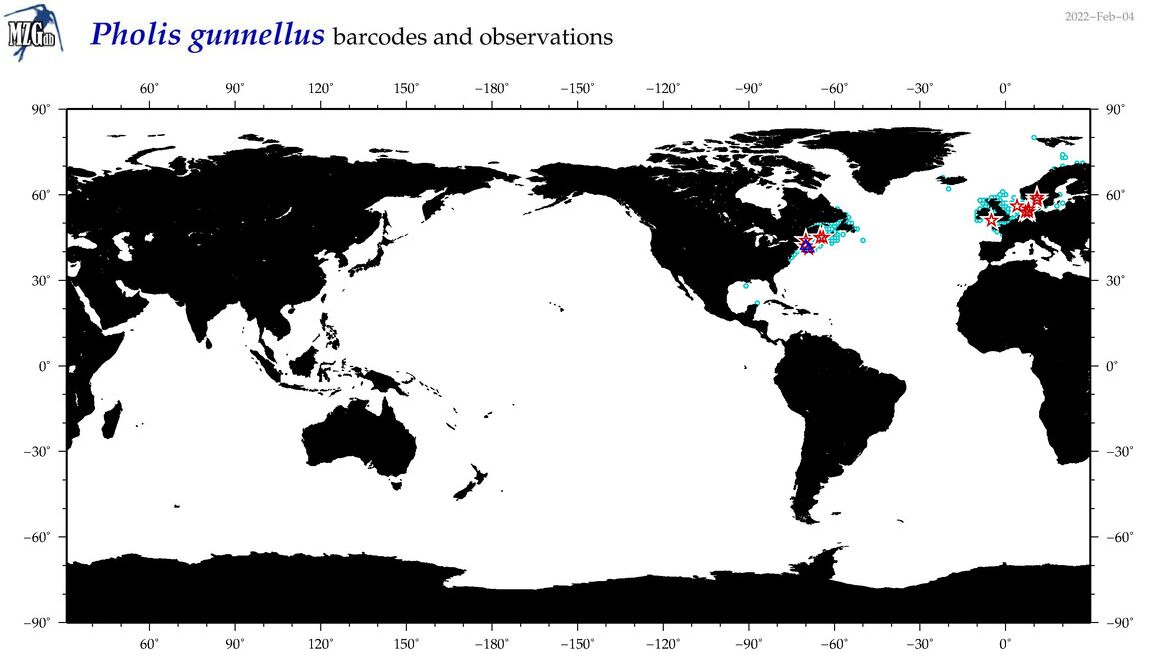
| Pholis gunnellus |
Species
(47) |
COI
12S
16S
18S
28S
|
COI = 21
12S = 6
16S = 5
18S = 0
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5001077
R:1:1:0:0 |
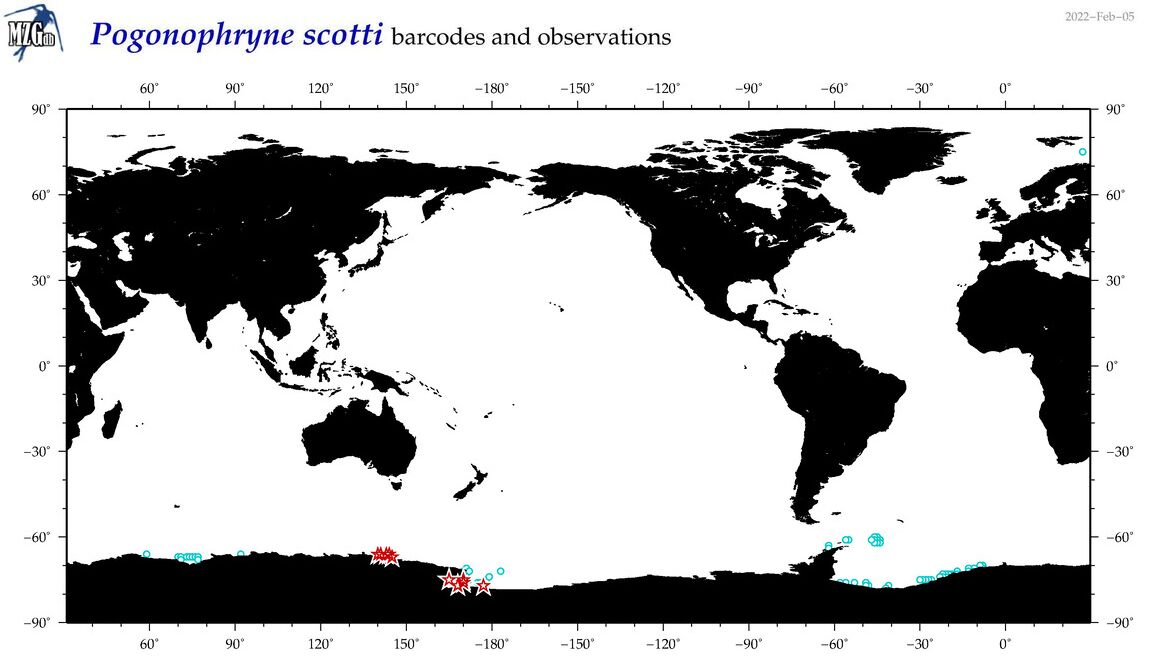
| Pogonophryne scotti |
Species
(48) |
COI
12S
16S
18S
28S
|
COI = 25
12S = 2
16S = 3
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5011524
R:1:0:0:0 |
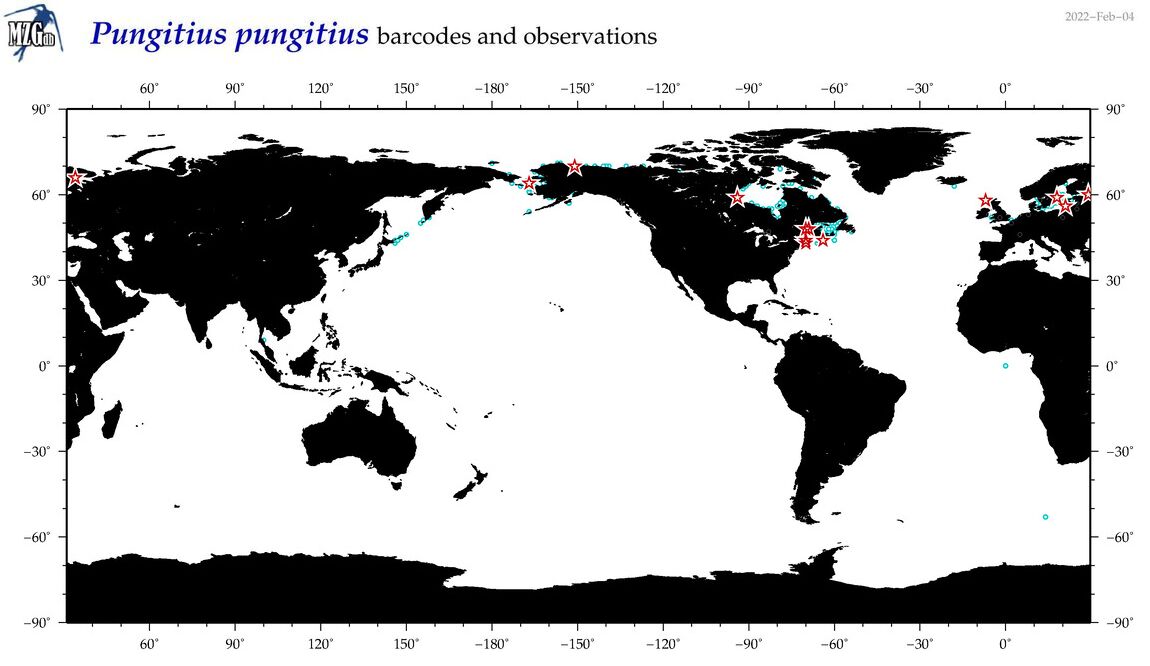
| Pungitius pungitius |
Species
(49) |
COI
12S
16S
18S
28S
|
COI = 102
12S = 18
16S = 39
18S = 0
28S = 0
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5001807
R:1:1:1:0 |
| Sebastes mentella |
Species
(50) |
COI
12S
16S
18S
28S
|
COI = 31
12S = 7
16S = 5
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
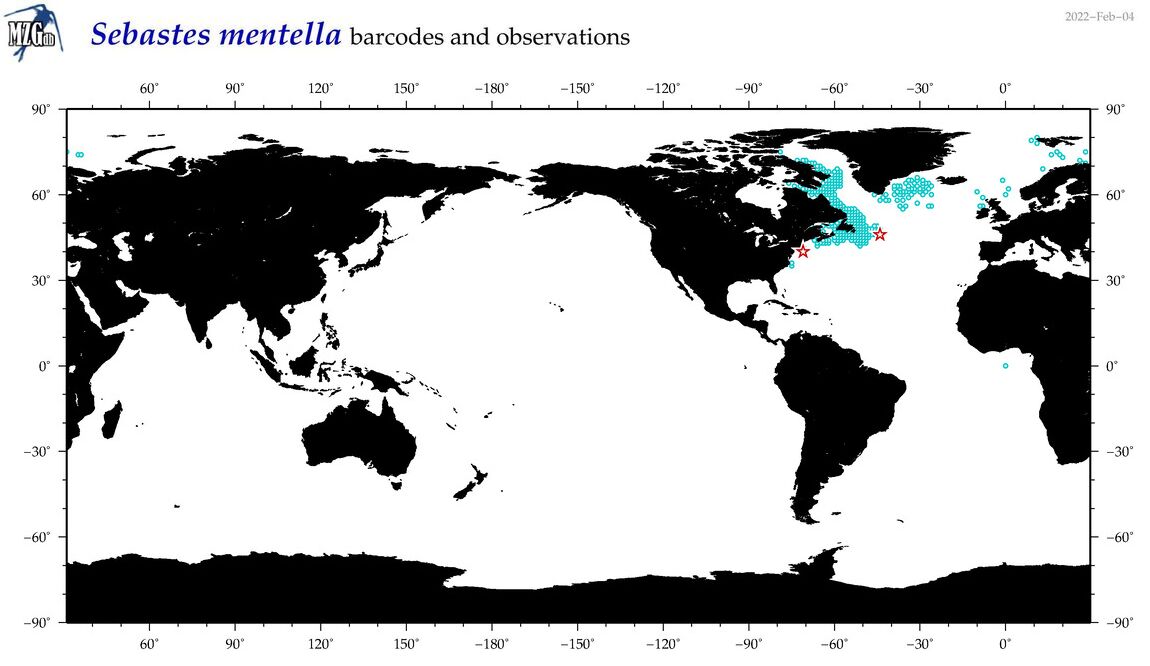 |
accepted
T5005493
R:1:0:0:0 |
| Sebastes norvegicus |
Species
(51) |
COI
12S
16S
18S
28S
|
COI = 20
12S = 5
16S = 4
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
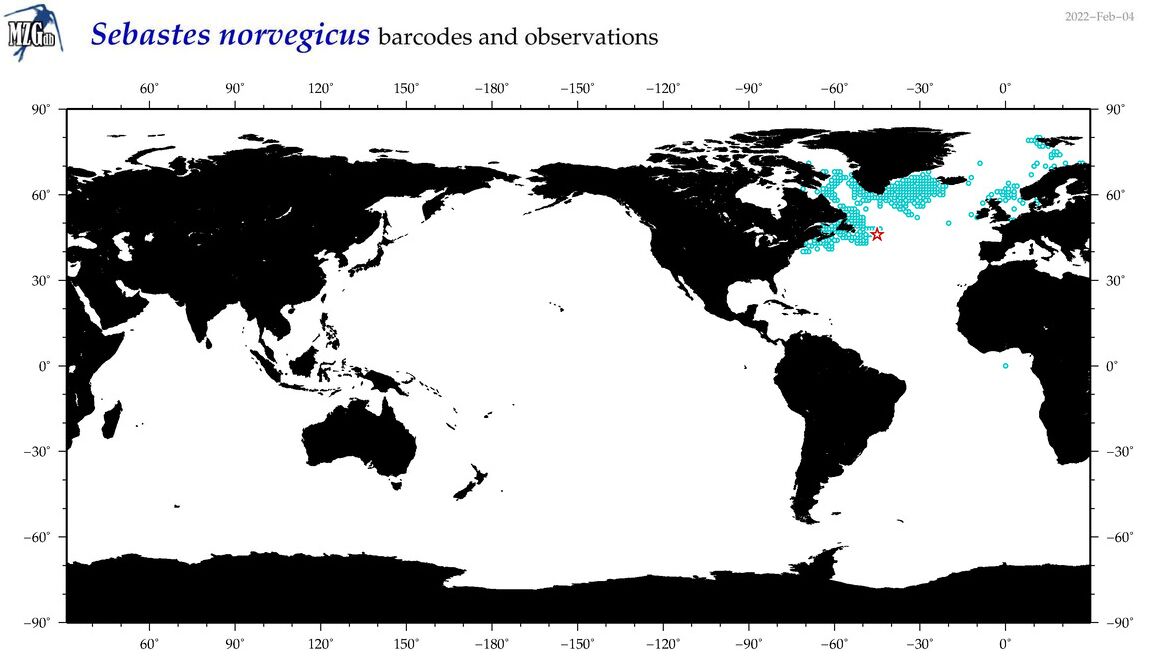 |
accepted
T5005500
R:1:0:0:0 |
| Sebastes viviparus |
Species
(52) |
COI
12S
16S
18S
28S
|
COI = 11
12S = 5
16S = 5
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
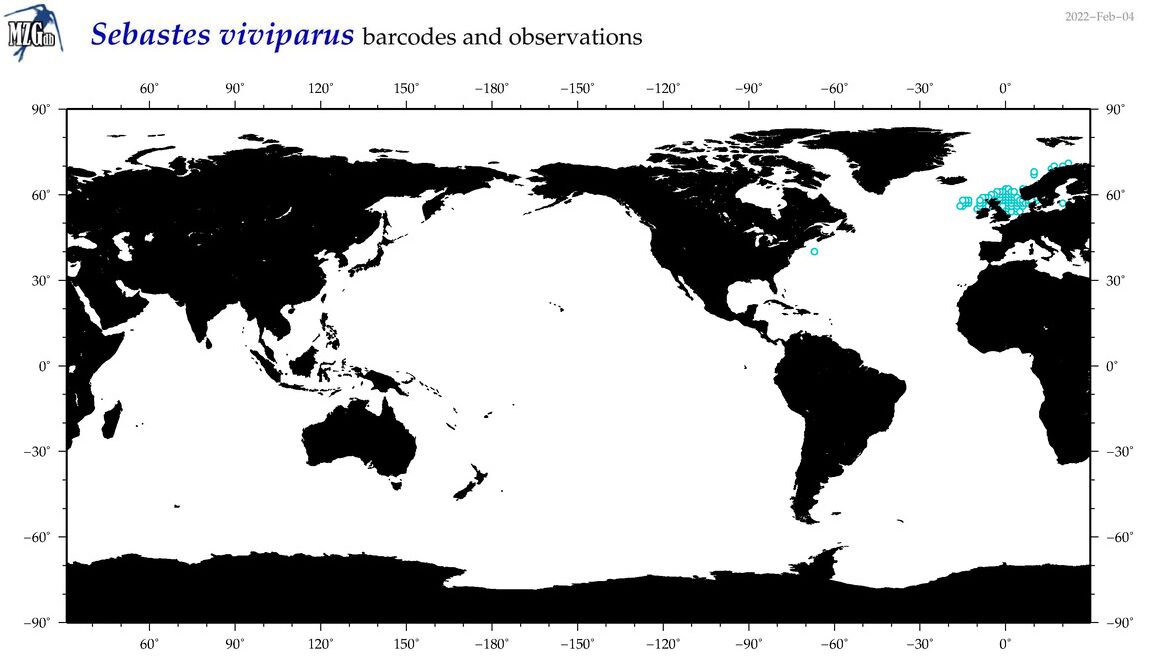 |
accepted
T5005535
R:1:0:0:0 |
| Taurulus bubalis |
Species
(53) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 3
16S = 6
18S = 0
28S = 2
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005723
R:1:1:0:0 |
| Triglops murrayi |
Species
(54) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 3
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
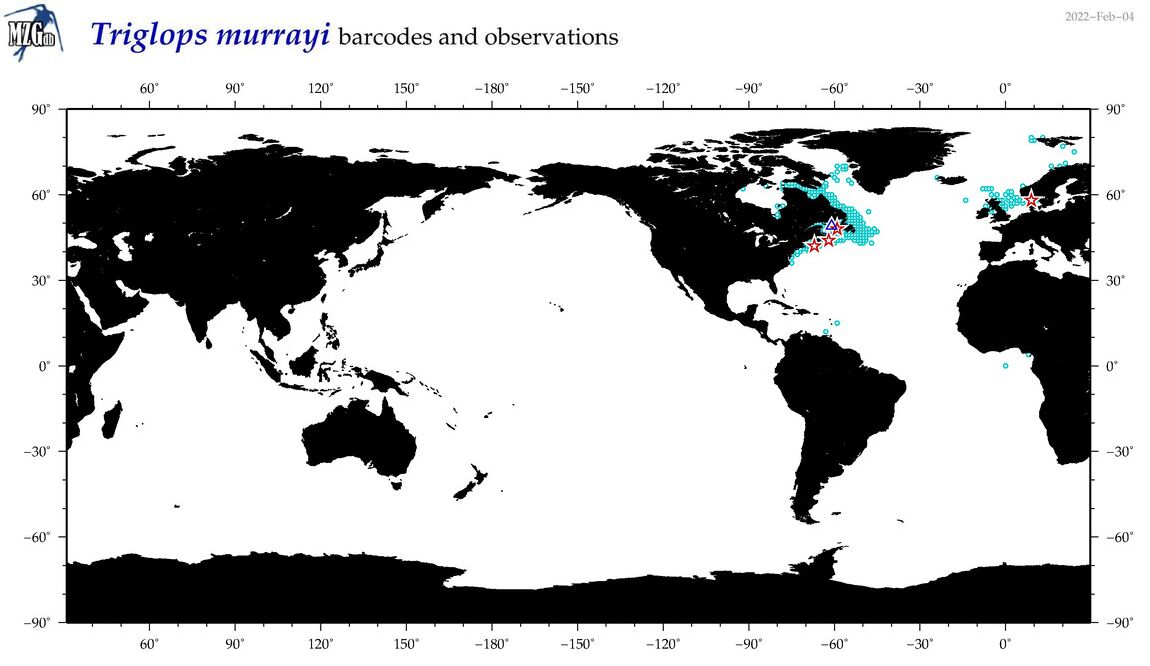 |
accepted
T5001373
R:1:0:0:0 |
| Triglops nybelini |
Species
(55) |
COI
12S
16S
18S
28S
|
COI = 31
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
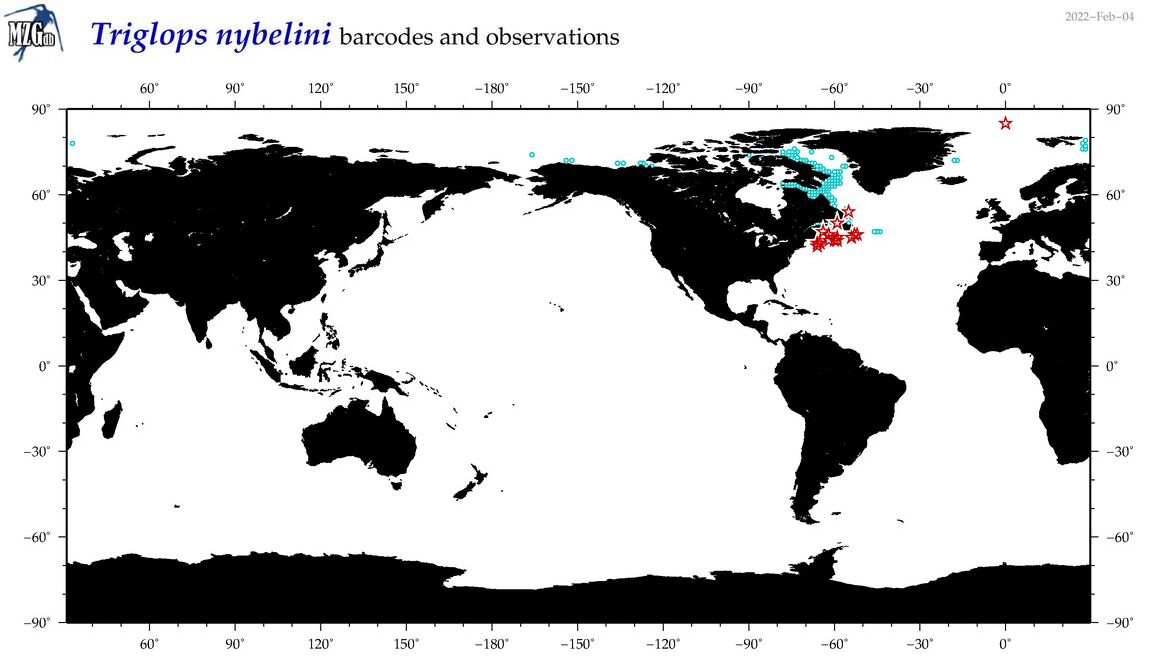 |
accepted
T5005797
R:1:0:0:0 |
| Triglops pingelii |
Species
(56) |
COI
12S
16S
18S
28S
|
COI = 34
12S = 7
16S = 5
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
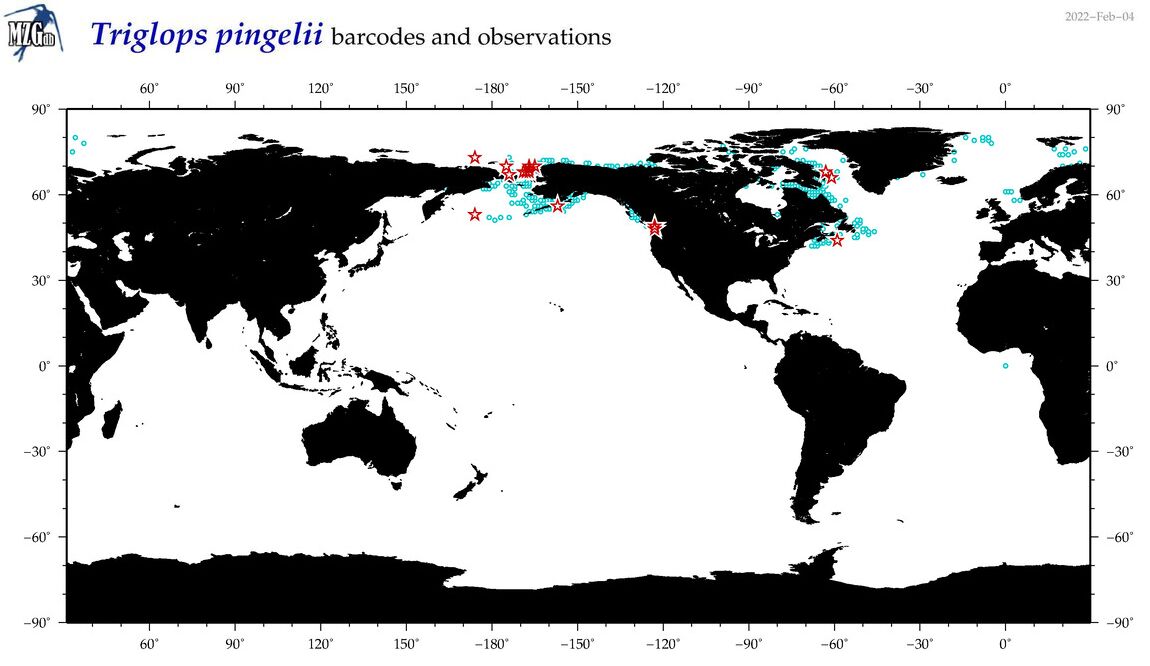 |
accepted
T5001374
R:1:0:0:0 |